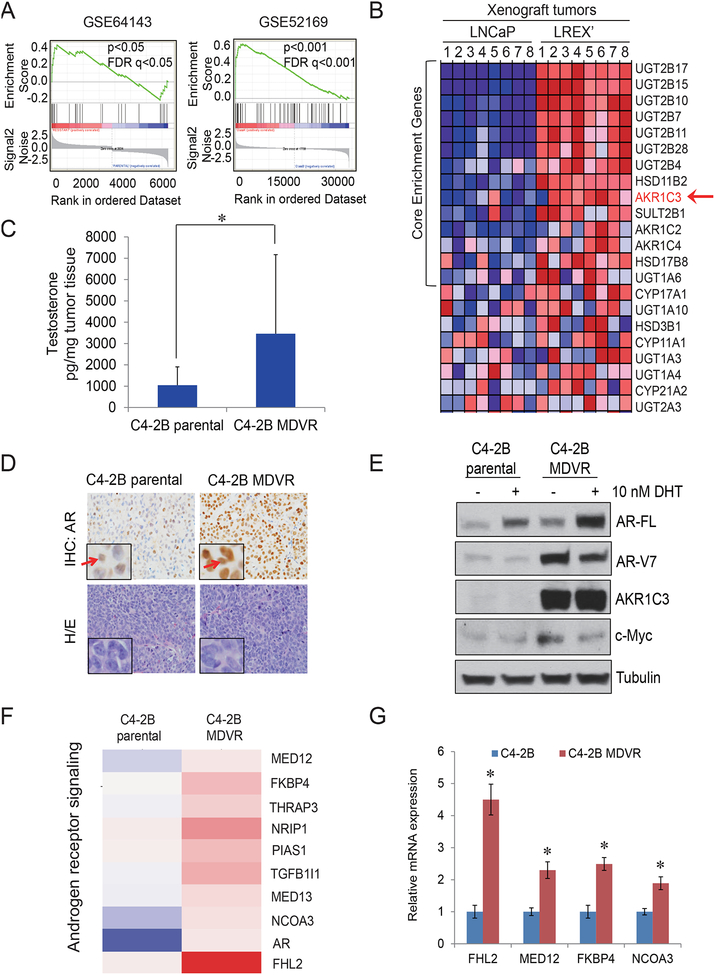
Figure 1. AR/AR-V7 signaling is reprogramed in C4-2B MDVR cells through AKR1C3 up regulation.
A. The steroid Hormone Biosynthesis pathway in two independent enzalutamide resistant models was analyzed by GSEA. GSE64143 is the microarray data set from C4–2B parental and C4–2B MDVR cells. GSE52619 is the mouse LNCaP/AR xenograft tumors derived from Enza resistant LREX’ cells compared with LNCaP xenograft tumors. B. Heatmap of Enza resistant xenograft tumors. C. C4–2B parental and C4–2B MDVR xenograft tumors were processed and the testosterone level was determined by LC-MS. D. AR staining was determined by IHC from C4–2B parental tumor and C4–2B MDVR tumor. E. C4–2B parental and C4–2B MDVR cells were cultured in CS-FBS condition and treated with 10nM DHT for 3 days, whole cell lysates was collected and subjected to western blot. F. Expression of transcripts encoding for androgen receptor signaling was analyzed. Genes that were regulated 1.3 fold between C4–2B parental cells and C4–2B MDVR cells were enriched and heat map was generated by Subio platform. G. Total RNA from C4–2B parental and C4–2B MDVR cells was extracted and the mRNA levels of FHL2, MED12, FKBP4 and NCOA3 were determined by qRT-PCR. AR-FL, AR-V7, AKR1C3, c-Myc were determined, tubulin was set up as control. * p<0.05