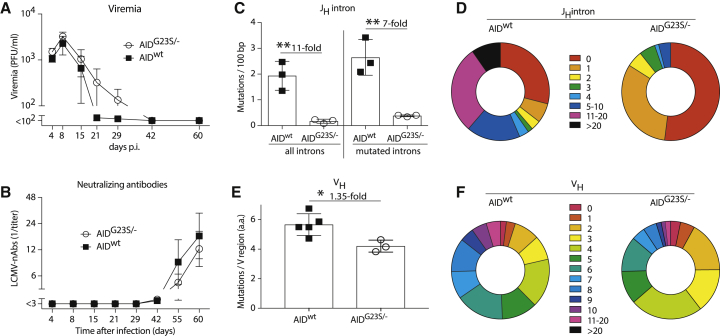
Figure 6.
Potent Selection of Hypermutated B Cells in Chronically Infected Mice
(A and B) We infected AIDG23S/- and WT control mice with rCl13 and measured viremia (A) and rCl13-neutralizing antibodies (B) over time.
(C and D) Sixty days after infection we sorted GC B cells (GL7+B220+) from the spleen for IgH locus sequencing. Number of nucleotide mutations in all JH introns (left) and in mutated JH introns (right, C). Distribution of JH intron mutation numbers (mutations per individual sequence) in AIDG23S/- and WT control mice (D). Results represent 74 and 62 sequences, respectively, from three mice per group.
(E and F) Number of amino acid mutations per sequenced VH region (E) and distribution of VH mutation numbers (F) in AIDG23S/− and AIDwt control mice. 1.3 ×106 – 1.5 × 106 sequences per mouse from three to five mice per group were analyzed.
Symbols in (A) and (B) represent means ± SEM. Bars in (C) and (E) represent means ± SD, with symbols showing individual mice. Donut plots represent the distribution of sequences with the indicated mutation numbers. n = 7 to 8 (A and B), n = 3 (C and D). N = 2 (A, B, E, and F), N = 1 (C and D). ∗p < 0.05, ∗∗p < 0.01 by unpaired two-tailed Student’s t test. The fold difference between groups is indicated (C and E).