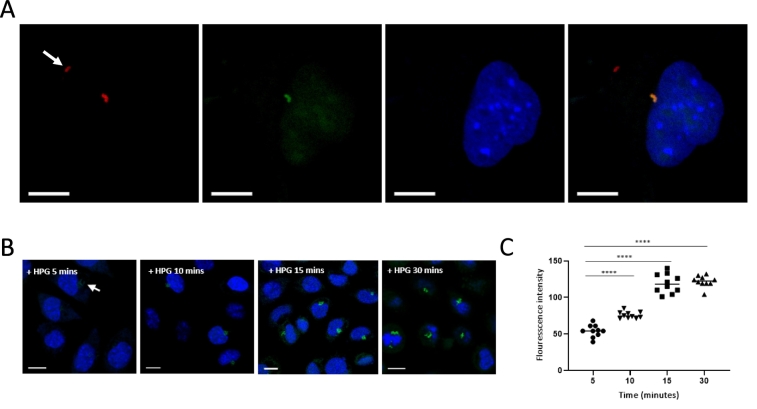
Fig. 4.
HPG can be used as a reporter of differential metabolic activity in bacterial subpopulations. A. Orientia tsutsugamushi were grown in L929 host cells with HPG for 30 mins in methionine-free media. After cell fixation HPG was reacted with an azide-conjugated fluorophore to reveal cells actively undergoing protein synthesis (green). Bacteria were counterstained in red (TSA56 antibody). This shows that metabolically inactive bacteria, located outside the host cell and indicated with a white arrow, cannot be labelled with HPG because they are not undergoing protein synthesis. B, C. The relative amount of HPG incorporated into bacteria can be estimated using fluorescence intensity measurements. Bacteria were grown in the presence of HPG for 5, 10, 15 and 30 min. The fluorescence intensity after 5 min incubation is lower than after 15 and 30 min as shown by confocal microscopy images (B) and fluorescence intensity measurements (C). Statistical significance was calculated using an unpaired t-test with GraphPad Prism software. ****p value ≤ .0001. The host cell nuclei are shown in blue (DAPI). Scale bar = 10 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)