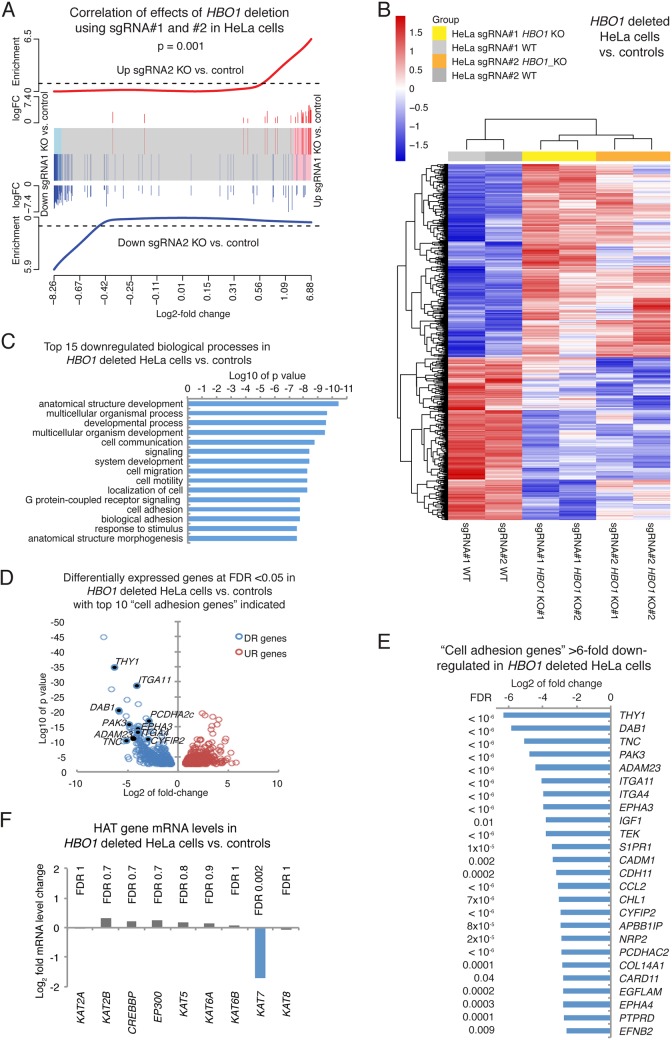
FIG 7.
Cell adhesion genes are downregulated in the absence of HBO1. Shown are RNA-Seq results for HeLa cells with an HBO1 deletion using sgRNA1 and sgRNA2 versus control cells (A to F) and combined analysis of sgRNA1 and sgRNA2 effects (B to F). (A) Barcode plot showing a high correlation between genes up- and downregulated as a consequence of the HBO1 deletion using sgRNA1 compared to sgRNA2 in HeLa cells. log FC, log fold change. (B) Heat map of genes differentially expressed between HBO1 knockout (KO) (guide 1, clones 1 and 2; guide 2, clones 1 and 2) and control HeLa cell (WT) clones induced for the expression of guide 1 or guide 2 in the absence of Cas9. (C) Biological process gene ontology terms enriched in genes downregulated in HBO1-null and control HeLa cells. (D) Volcano plot displaying log10 P values over log2 fold expression differences between HBO1-null and control HeLa cells, with downregulated (DR) “cell adhesion genes” indicated. UR, upregulated. (E) Cell adhesion genes >6-fold downregulated in HBO1-null and control HeLa cells. Two clonal control cell lines (one each for sgRNA1 and -2) and 4 clonal HBO1-null cell lines (two each for sgRNA1 and -2) were used. Data were analyzed by RNA sequencing as described in Materials and Methods. Genes differentially expressed with an FDR of <0.05 were considered significant. Related data on 293T and HeLa cells are displayed in Fig. S6 and S7 in the supplemental material. All data are provided in Tables S4 and S5. (F) Histone acetyltransferase (HAT) gene expression, apart from HBO1, is not changed.