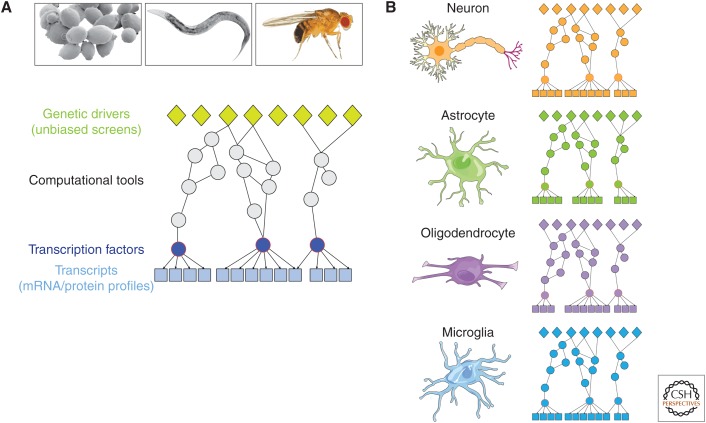
Figure 2.
Building a global view of cellular response to proteotoxicity. (A) Bioinformatic methods can be applied to bridge data sets from genome-wide genetic screens and transcription profiles, thereby integrating genetic drivers and cellular response to proteotoxicity, and augmenting the list of molecular candidates implicated in the cytotoxic mechanism. Network-building algorithms can identify “hidden nodes” that are not recovered in individual screens. Such approaches are currently feasible in model organisms where multiple “omics” data sets are already available, but can one day be applied to human disease-relevant cell types (e.g., human neurons) as genome-wide genetic and molecular data sets become available. (B) Different cell types (e.g., neurons, astrocytes, oligodendrocytes, microglia) likely exhibit different molecular network profiles due to differential gene expression, so within proteotoxicity models, it will be important to obtain cell-specific data and build cell-type-specific networks. mRNA, messenger RNA.