Figure 4. RNase J1 disassembles stalled transcription complexes (TC).
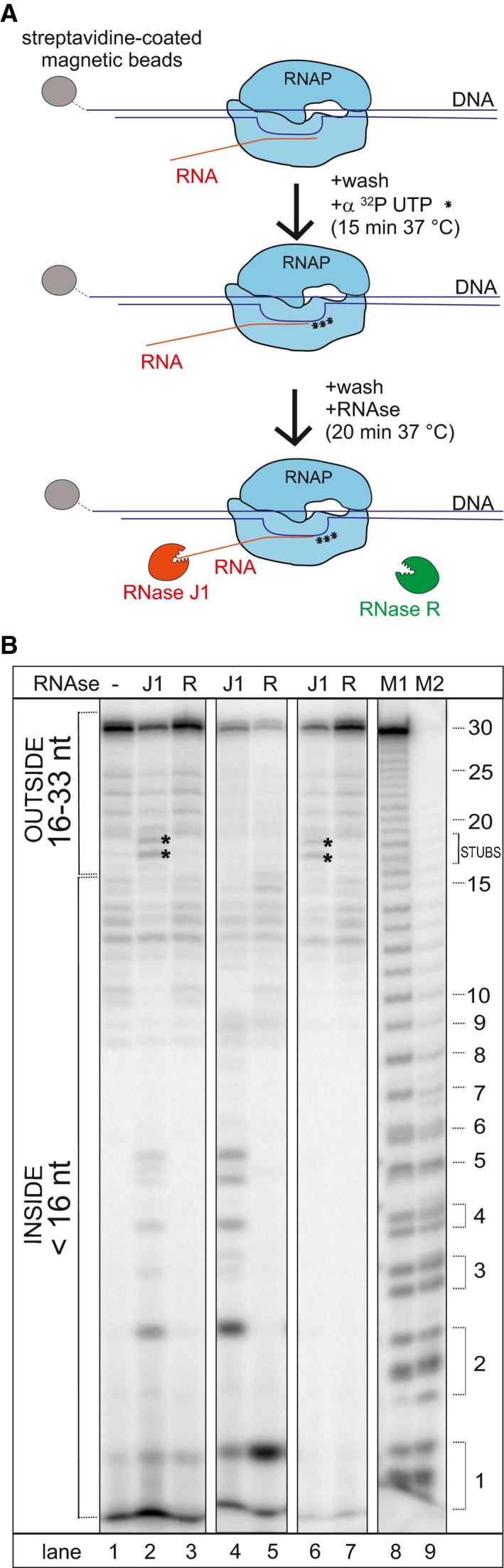
- Schematic representation of the flow of the experiment. TCs were assembled on DNA‐RNA scaffolds (DNA was biotinylated); RNA was labeled at 3′ with radioactive UTPs (asterisks). The RNA length (including label) was 33 nt.
- Representative primary data—polyacrylamide gel (the experiment was performed 3× with the same results). Lane 1, the full‐length (33 nt) labeled RNA; lane 2, the same as lane 1 but it included also incubation with RNase J1 (J1), 17–18 nt long fragments (RNase J1 stopped by RNAP; indicated with asterisks) and < 5 nt fragments (RNA released from TC into buffer—indicative of TC disassembly) are shown; lane 3, the same as lane 1 but included also incubation with RNase R (R); lanes 4 and 5, TCs were denatured by heat prior to RNase addition to demonstrate the activity and cleavage patterns of both enzymes; lanes 6 and 7, the same as lanes 2 and 3 but the buffer was washed off (TCs were retained by streptavidin beads) to demonstrate which RNA fragments were associated with TC; lanes 8 and 9 (M1, M2) Mw marker generated by treating the 30 nt RNA with alkali and formamide (M1—4‐min treatment, M2—7‐min treatment). As reported in Costanzo et al (2016) (and references therein), the cleavage by alkali or formamide leaves the phosphate group of the attacked phosphodiester bond bound at 3′, initially in the 2′,3′ cyclic form (upper band in the band couples). This successively opens (lower band in the band couples) yielding a double‐banded pattern for short oligoribonucleotides.
Source data are available online for this figure.
