Figure 1.
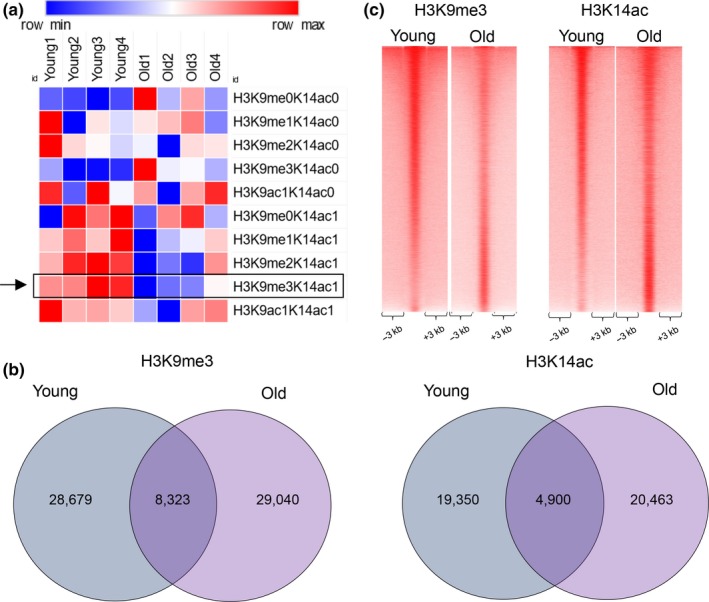
New bivalent chromatin state identified by proteomic analysis in mammalian liver. (a) Heatmap showing the relative intensities of histone marks determined by targeted mass spectrometry of histone tails in young and old livers (values are normalized to row's median). A bivalent H3K9me3/H3K14ac mark (marked with a rectangle and an arrow) was identified in both young and old livers and was quantitatively reduced in old livers. (b) Venn diagram showing the results of genome‐wide location analysis for H3K9me3 (left panel) and H3K14ac (right panel) marks in young and old liver. For H3K9me3, Peak‐Seq identified 28,679 sites in young liver and 29,030 in old liver, with 8,323 in the overlap. For H3K14ac, 19,350 peaks in young liver and 20,463 peaks in old liver were called, with 4,900 in the overlap. (c) Heatmaps comparing H3K9me3 (left panel) and H3K14ac (right panel) ChIP‐Seq coverage in young and old livers. Overall, there was more H3K9me3 but less H3K14ac signal in young livers as compared to the old. Reads were merged from two replicates for H3K9me3 and H3K14ac ChIP‐Seq data in both conditions. Data for H3K9me3 were accessed from our previous study (Whitton et al., 2018)
