Figure 5.
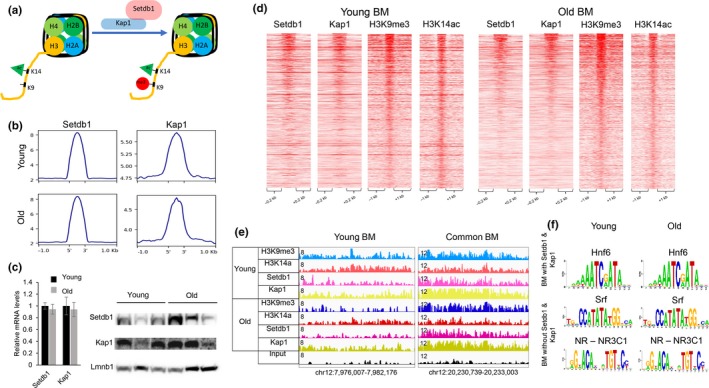
Setdb1/Kap1 complex associates with bivalent regions. (a) Model implicating histone methyltransferase, Setdb1, in establishing regions with bivalent mark. Setdb1, in complex with Kap1, methylates H3K9 by recognizing H3K14 acetylation, creating a new dually marked region. (b) Profile plot generated by deeptools showing Setdb1 (left panel) and Kap1 (right panel) signal at Setdb1‐bound regions in young (top panel) and old (bottom panel) livers. Average number of reads per bin (25 bp) is shown on y‐axis. Reads were merged from two biological replicates in each condition. (c) Relative mRNA levels (n = 4, left panel) of Setdb1 and Kap1 by quantitative RT‐PCR. Western blot analysis (n = 3, right panel) of protein nuclear extracts with antibodies to Setdb1, Kap1, and Lmnb1 (loading control) in young and old livers. mRNA and protein expression levels of Setdb1 and Kap1 did not change significantly. Gapdh was used as normalizing control in quantitative RT‐PCR. (d) Heatmaps showing Setdb1, Kap1, H3K9me3, and H3K14ac ChIP‐Seq signal at bivalent regions of young (left panel) and old (right panel) livers. A subset of regions with bivalent mark is bound by Setdb1 and Kap1 in both young and old livers. (e) Examples of genomic regions showing bivalent regions bound by Setdb1/Kap1 complex in young (chr12:7,976,007–7,982,176) and common to young and old livers (chr12:20,230,739–20,233,003) livers. Magnitude of the ChIP‐Seq signal is shown on y‐axis. (f) Overrepresented motifs in the bivalent regions with and without Setdb1/Kap1 binding in young and old livers, identified by PscanChIP. Hnf6 motifs were enriched in bivalent regions occupied by Setdb1/Kap1 in both young and old livers (p‐values < 1.4E‐34 and < 3.8E‐28, respectively). In contrast, motifs in dually marked regions without Setdb1/Kap1 occupancy included Srf in both young and old livers (p‐values < 5.5E‐6 and 0.0088, respectively) and several nuclear receptors (NR3C1, young NR3C1, p‐values«.0002 and .0006, respectively). ChIP‐Seq data for H3K9me3 are from our previous study (Whitton et al., 2018)
