Abstract
Background/Purpose
Uropathogenic E. coli (UPEC) is the main cause of urinary tract infection (UTI) and it is known that pregnant women have a higher risk for UTI. UPEC has a variety of virulence and antibiotic resistance factors that facilitate its pathogenic success and it is crucial to know which are the susceptibility patterns, Extended-Spectrum-β-Lactamase (ESBL) production, virulence genes, pathogenicity islands (PAI), phylogenetic groups and serotypes among strains isolated from pregnant and non-pregnant women.
Methods
One hundred fifty UPEC strains were isolated from pregnant and non-pregnant women from two different Mexican states (Sonora and Puebla). Strains were analyzed using the Kirby-Bauer method for the determination of antibiotic susceptibility and ESBL. Virulence genes, PAIs and phylogenetic groups were determined using a multiplex PCR. Strains were serotyped by an agglutination assay. Blood agar and CAS agar were used for phenotypic assays.
Results
92.7% of UPEC strains showed multidrug-resistant (MDR), 6.7% extremely-resistant (XDR) and 0.6% pandrug-resistant (PDR). The highest resistance was determined to be for β-lactam antibiotics (>72% in both states) and 44.5% of the UPEC strains were ESBL+. The predominant virulence genes found were fimH (100%), iucD (85%) and iha (60%). The strains isolated from pregnant women from Puebla presented a large percentage of genes associated with upper urinary tract infections. PAIs were found in 51% and 68% of the strains from Sonora and Puebla, respectively. All the strains were siderophores producers and 41.5% produced hemolysis. The serotypes found were diverse and belonged to phylogroups A, B2 and C.
Conclusion
The UPEC strains from this study are MDR with tendency to XDR or PDR, they can cause upper UTIs and are serotypically and phylogenetically diverse, which supports the need to develop new strategies for UTI treatment in pregnant and non-pregnant Mexican women.
Keywords: UPEC in pregnancy, antibiotic resistance, virulence profile, phylogenetic groups, multiplex PCR, serotype
Introduction
Urinary Tract Infections (UTI) are defined as the presence of microorganisms in any type of sterile organ of the urinary tract. UTIs are one of the most common disease worldwide, affecting 150 million people every year, with women being the most affected group. During 2017, 4,054,073 cases of UTI were diagnosed in Mexico, being females the most vulnerable group with 82.1% of the reported cases.1 During pregnancy, the risk of acquiring an UTI triplicates due to the wide range of anatomical, functional and hormonal changes inherent to the gestational stage being considered a risk factor for the health of both the mother and the fetus.2–4 UTIs are classified based on their anatomical location in upper (pyelonephritis) or lower (cystitis). In both cases, uropathogenic Escherichia coli (UPEC) is the main etiologic agent.5–8
UPEC belongs to the Extraintestinal Pathogenic E. coli group (EXPEC) and it is associated with a subset of serogroups and serotypes (O1:H4, O1:H6, O1:H7, O1:H−, O2:H1, O2:H4, O4:H5, O6:H1, O7:H4, O7:H6, O7:H-, O18ac:H7, O18ac:H-, O22:H1, O25:H1, O75:H5 & O75:H7) and with the B2 or D phylogenetic groups.9–12 UPEC possess a wide number of virulence and resistant determinants that allows it to successfully colonize the urinary tract and cause disease.13–16 Within the most frequent virulence factors reported in UPEC isolates are those involve in adherence (fimbrial and afimbrial adhesins) such as FimH, PapG; iron acquisition systems such as Aerobactin (which belong to hydroxamate family), Salmochelin, Enterochelin or Yersiniabactin; toxigenic proteins (HlyA, CNF-1, Sat, Vat) and motility (flagellum).13,16–18 Usually, both virulence and antibiotic resistant factors, are located in mobile elements such as plasmids or genomic islands, which are highly interchangeable between bacterial strains.9,19
In Mexico, UTI treatment is generally empirical. The drugs most commonly used in uncomplicated UTI are second and third-generation cephalosporins, trimethoprim with sulfamethoxazole, nitrofurantoin and fosfomycin.20–23 However, for pregnant women the treatment is different, due to the pharmacokinetics and pharmacodynamics alterations that can occur during pregnancy; therefore, the number of antibiotics available for treatment is significantly reduced.22–24 One disadvantage of empirical treatment is the high risk of generating antibiotic resistance. This phenomena currently represents one of the main-health problems worldwide, since there are fewer drugs capable of eliminating resistant and multidrug resistant (MDR) microorganisms.25 In this sense, there are many mechanisms that confer antibiotic resistance in bacteria, one of the most studied is the production of Extended-Spectrum β-lactamases (ESBL) such as TEM, OXA, and SHV that are responsible of the resistance to broad-spectrum penicillins, cephalosporins and monobactams. To date, more than 350-different ESBL have been described and mainly are encoded in plasmids.26–28 Some investigations indicated that depending on enzyme substrate, the strains can be classified as ESBL producers or carbapenemases producers.29,30 This is important because carbapenems are in the list of last resource antibiotics for treatment.
Despite the high incidence of UPEC causing UTI in Mexico, there are few researchers focusing on UPEC pathogenesis and characterization of their determinants. Therefore, the objective of this work was to analyze the virulence features, resistance profiles, serotypes and phylogenetic groups of uropathogenic E. coli strains isolated from pregnant and non-pregnant Mexican women from two different geographical areas.
Materials and Methods
Biological Samples
One hundred fifty E. coli clinical isolates were obtained from urine samples from same number of pregnant and non-pregnant women, in fertile age, from two-Mexican states, Sonora and Puebla, during the period April 2017 to December 2018. Fifty strains were from Puebla and 100 from Sonora. Half of the strains from each city were obtained from pregnant women. The urine samples were obtained in a sterile container by midstream clean-catch and the strains were identified by conventional biochemical test. The Sonora’s strains were donated by the clinical analysis laboratory “UNILABS” of the Universidad de Sonora, in Caborca city and Puebla’s strains were donated by the Nephrourological Research Laboratory of the Faculty of Chemical Sciences of the BUAP. All the E. coli strains were sent to the Centro de Investigacion en Ciencias Microbiologicas under optimal conditions as dictated by the guide of regulations regarding the transport of infectious substances (WHO/HSE/GCR/2012.12) and NOM-051-SCT2/2011)31 for further analysis including selective medium culture (MacConkey agar), re-identification by conventional biochemical test and confirmed as E. coli using GNI card (Gram-negative identification) of the VITEK 2 System (BioMérieux). The reference uropathogenic E. coli strains CFT073 and GAG1 were kindly provided by Dr. Jose Molina and the E. coli strains J96 were kindly provided by Dr. Juán Xicohtencatl-Cortes.
Serotyping
The strains were serotyped by agglutination assay using 96-well microtiter plates and rabbit serum obtained against 187 somatic antigens (O) and 53 flagellar antigens (H) for E. coli.32
Chromosomic and Plasmidic DNA Extraction
The DNA extraction was done by alkaline lysis according to established protocols by the Molecular cloning A Laboratory Manual, 2012.33
Determination of Antibiotic Susceptibility Profiles and ESBL
The antibiogram was done using the Kirby-Bauer disk diffusion method, where 24 antibiotics (Supplementary material 1) were analyzed following the guidelines of the Institute of Clinical and Laboratory Standards. E. coli ATCC25922 and P. aeruginosa ATCC27853 were used as controls.34 ESBL production was determined by the double-disk diffusion method, following the criteria established by the CLSI. The antibiotics included in the assay were cefotaxime, ceftazidime, ceftriaxone, cefepime, and aztreonam. Amoxicillin with clavulanic acid was used as an inhibitor. Magiorakos’ criteria were followed to classify the strains as non-multidrug resistant (NMDR); multidrug-resistant (MDR), extremely resistant (XDR), or pandrug resistant (PDR).35
Virulence Associated Genes
We evaluated 13 virulence genes associated to 9 virulence factors: Type 1 pilus (fimH), flagellin (fliC), secreted autotransporter toxin (Sat: satA and satP), aerobactin (iucD), the bifunctional siderophore receptor/adhesin (iha), vacuolating autotransporter toxin (Vat: vatA and vatP), type P pilus (papA, papGII and papGIII), Hemolysin A (hlyA) and cytotoxic necrotizing factor (cnf-1). Three multiplex PCR (mPCR1, mPCR2 and mPCR3) and one individual PCR (PCRcnf-1) (Supplementary material 2) were used. All PCR products were analyzed in a 1% agarose gel by electrophoresis following a 0.084 mM ethidium bromide staining. In the case of Sat, Vat and P pilus, only the strains with all associated genes were considered as positive. Negative results were confirmed by individual PCR. CFT073, GAG-1, O59I and J96 were used as positive controls.
Determination of Pathogenicity Islands (PAIs)
The protocol established by Sabate in 200636 was used to identify PAIs I and II of E. coli CFT073 and E. coli J96 (Supplementary material 2).
Phylogenetic Groups
The phylogenetic group identification was carried out according to the second scheme by Clermont in 2013.37
Iron Acquisition Phenotype
All strains were seeded in previously perforated CAS agar and sealed with bacteriological agar for detection of iron acquisition phenotype following protocols already established.38–40 In brief, the strains were inoculated in 1 mL of LB medium for 18 h, the culture was adjusted to an optical density to 600 nm of 0.1 (OD600nm 0.1) using LB medium deferred with 3% hydroxyquinoline and chloroform. It was incubated until exponential phase and 25 μL were plated in CAS agar previously perforated and sealed with bacteriological agar, the plates were incubated for 32 h and a color change characteristic of the presence of siderophores in the culture medium was observed.
Hemolysis Phenotype
Twenty microliters of a preculture in LB medium incubated to 37°C for 24 h of each strain were inoculated in previously perforated blood agar with 5% sheep blood, and sealed with bacteriological agar, they were incubated for 24 h and the presence of hemolysis was observed around the inoculum. This test was carried out for all strains obtained in this study.
Disposal of Microorganisms and Reagents
It was carried out in accordance with the Official Mexican Standard NOM-087-SEMARNAT-SSA1-2002 for RPBI and NOM-052-SEMARNAT-2005 for CRETI waste.
Statistical Analysis
The results were analyzed using two samples test and Fisher test using the Minitab 18, Statistix 10 trial and GraphPad Prims 6 software. The level of significance was set at a p value ≤0.05.
Ethics
All the isolates were collected during routine sampling. The patients were informed of the objective of the project and they signed a written informed consent. The patient´s data were maintained under anonymity. Approval by an ethics committee was not required because all the strains used in this study were donated by two-clinical laboratories located in Sonora and Puebla, respectively.
Results
Clinical Isolates of E. coli Obtained from Urine of Pregnant and Non-Pregnant Women in Sonora and Puebla are Multidrug-Resistant
Antibiotic resistance is one of the most important problems in health settings worldwide because of the decreasing number of effective antibiotics. In the present study, we analyzed the susceptibility of the isolated UPEC strains to 24-different types of antibiotics. The predominant resistance in all strains from both Mexican states and from each group was against β-lactamic antibiotics. The strains showed also a high-resistance profile against nitrofurantoin, trimethoprim/sulfamethoxazole, amikacin and gentamicin (Table 1). Resistance to fluoroquinolones was also observed in all study groups. On the other hand, ertapenem, netilmicin, and fosfomycin were the most effective antibiotics among the 24 antibiotics evaluated. We did not find statistical differences in the mean of resistance to the 24 antibiotics tested into each study group between geographical areas (p ≥ 0.05) (Supplementary material 3). However, the specific resistance for each antibiotic between the two different geographical areas was significant. The strains from pregnant women in Puebla presented significant differences in resistance for amikacin (p=0.001), cefuroxime (p=0.025) and fosfomycin (p=0.037), and the strains from Sonora for levofloxacin (p=0.049). In the case of non-pregnant women, there was only a significant difference in the higher antibiotic resistance to netilmicin (p=0.037) of strains from Puebla compared to the ones from Sonora (Supplementary material 3). Between study groups from Sonora, a statistical difference was observed in the resistance to levofloxacin and tetracycline in pregnant women and cefuroxime, cefotaxime, ceftriaxone, amoxicillin/clavulanic acid and nitrofurantoin in non-pregnant women. While in Puebla, there was statistical significance only in the resistance to ertapenem in strains isolated from not pregnant women (Table 1). According to the Magiorakos scheme, 92.7% of strains were multidrug-resistant (MDR); however, we also found 10 (6.7%) XDR and one (0.6%) PDR strains (Table 2). The PDR strain was resistant to 24 antibiotics tested in this study.
Table 1.
Antibiotic Resistance of E. coli Strains Isolates from Urine of Pregnant and Non-Pregnant Women from Sonora and Puebla, Mexico
| Antibioticsa | Sonora (n=100) pb = 0.840 | Puebla (n=50) pb = 0.946 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pregnant n=50 n (%) | Average %R | Non-Pregnant n=50 n (%) | Average %R | pc value | Pregnant n=25 n (%) | Average %R | Non-Pregnant n=25 n (%) | Average %R | pc value | |
| AMK | 31 (62) | 38 | 32 (64) | 37.3 | 1 | 24 (96) | 58.7 | 19 (76) | 45.3 | 0.098 |
| GM | 21 (42) | 22 (44) | 1 | 14 (56) | 10 (40) | 0.396 | ||||
| NET | 5 (20) | 2 (4) | 0.436 | 4 (24) | 5 (20) | 1 | ||||
| AMP | 50 (100) | 72.9 | 50 (100) | 78.9 | – | 100 (25) | 82.2 | 25 (100) | 77.3 | – |
| CF | 48 (96) | 49 (98) | 1 | 24 (96) | 25 (100) | 1 | ||||
| CFX | 41 (82) | 50 (100) | 0.002 | 25 (100) | 23 (92) | 0.489 | ||||
| CTX | 35 (70) | 47 (94) | 0.003 | 20 (80) | 20 (80) | 1 | ||||
| CFZ | 37 (74) | 33 (66) | 0.513 | 21 (84) | 19 (76) | 0.725 | ||||
| CRO | 41 (82) | 50 (100) | 0.002 | 24 (96) | 24 (96) | 1 | ||||
| FEP | 19 (38) | 14 (28) | 0.395 | 8 (32) | 6 (24) | 0.753 | ||||
| ATM | 20 (40) | 15 (30) | 0.401 | 15 (60) | 11 (44) | 0.396 | ||||
| AMC | 37 (74) | 47 (94) | 0.012 | 23 (92) | 22 (84) | 0.667 | ||||
| NA | 37 (74) | 60 | 37 (74) | 44.8 | 1 | 17 (68) | 44 | 18 (72) | 56 | 1 |
| CIP | 29 (58) | 22 (44) | 0.229 | 10 (40) | 13 (52) | 0.570 | ||||
| OFX | 27 (54) | 18 (36) | 0.107 | 12 (48) | 14 (56) | 0.777 | ||||
| NOR | 28 (56) | 19 (38) | 0.108 | 13 (32) | 14 (56) | 0.153 | ||||
| LVX | 29 (58) | 16 (32) | 0.015 | 13 (32) | 11 (44) | 0.560 | ||||
| NF | 20 (40) | 40 | 41 (82) | 82 | <0.001 | 16 (64) | 64 | 17 (68) | 68 | 1 |
| TSX | 32 (64) | 64 | 35 (70) | 70 | 0.670 | 11 (44) | 44 | 17 (68) | 68 | 0.153 |
| FOS | 2 (4) | 4 | 2 (4) | 4 | 1 | 5 (20) | 20 | 1 (4) | 4 | 0.189 |
| C | 30 (60) | 70 | 25 (50) | 50 | 0.421 | 9 (36) | 36 | 11 (44) | 44 | 0.773 |
| TE | 35 (70) | 70 | 29 (58) | 58 | 0.032 | 13 (52) | 52 | 18 (72) | 72 | 0.243 |
| ETP | 6 (12) | 12 | 4 (8) | 8 | 0.740 | 5 (20) | 20 | 0 (0) | 0 | 0.050 |
Notes: pb, two samples t-test; pc, Fisher test exact; –, the value of p could not be obtained; n, number of strains; %, percentage; R, resistance; statistically significant values are in bold.
Abbreviations: AMK, amikacine; GM, gentamicin; NET, netilmicine; AMP, ampicillin; CF, cephalotin; CFX, cefuroxime; CTX, cephotaxime; CFZ, cefazolin; CRO, cephtriaxone; FEP, cephepime; ATM, Aztreonam; AMC, clavulanic acid-ampicillin; NA, nalidixic acid; CIP, ciprofloxacin; OFX, ofloxacin; NOR, norfloxacin; LVX, levofloxacine; NF, nitrofurantoin; TSX, trimethoprim/sulfamethoxazole; FOS, fosfomycin; C, chloramphenicol; TE, tetracycline; ETP, Ertapenem.
Table 2.
Antibiotic Profile Classification of E. coli Isolates from Urine of Women from Sonora and Puebla, Mexico in Relation to Pregnant and Non-Pregnant Conditions
| City | Condition | Classification by Drug-Resistance n=150 (100%) | |||
|---|---|---|---|---|---|
| NMDR n (%) | MDR n (%) | XDR n (%) | PDR n (%) | ||
| 0 (0) | 139 (92.7) | 10 (6.7) | 1 (0.6) | ||
| Puebla | Pregnant (n=25) | 0 | 22 (88) | 2 (8) | 1 (4) |
| Non-pregnant (n=25) | 0 | 24 (96) | 1 (4) | 0 | |
| Sonora | Pregnant (n=50) | 0 | 47 (94) | 3 (6) | 0 |
| Non-pregnant (n=50) | 0 | 46 (92) | 4 (8) | 0 | |
Abbreviations: NMDR, Non-multidrug resistant; MDR, Multidrug-resistance; XDR, extensively multidrug-resistance; PDR, pandrug-resistance.
Puebla and Sonora’s E. coli Strains from Urine Samples are ESBL Producers
The ESBL phenotype was found in 67 (44.7%) of the total of strains analyzed (n=150). Twenty-two (44%) strains isolated from Puebla were ESBL positives, 17 from pregnant women and 5 from not pregnant women. We found in this group that 90% (n=20) of the UPEC strains with positive β- lactamase phenotype were classified as ESBL producers and 10% (n=2) are probable carbapenemases producers, these last ones come from pregnant women (Supplementary material 4.1). In Sonora, the β-lactamase phenotype was observed in 45 (45%) of the isolated strains, 22 were from pregnant women and 23 from non-pregnant women. Also 91% (n=41) of the E. coli strains with positive β- lactamase phenotype were classified as ESBL producers and 9% (n=4) as probable carbapenemases producers (Supplementary material 4.2 and Supplementary material 4.3). To find whether there was a relationship between the antibiotic resistance and the β-lactamase production, we compare the prevalence of resistance between β-lactamase producers’ strains and non-β-lactamase producers. We observed that, in general, the strains with β-lactamase phenotype, except in the case of non-pregnant women from Puebla, they had a higher prevalence of antibiotic resistance than those from non-β-lactamase producers, but only showed statistical significance (p ≤ 0.05) for TSX, FEP, AMC and ATM in Sonora and NOR, LVX and FEP in strains from pregnant women from Puebla (Supplementary material 5). These results could indicate a phenomenon of co-selection of resistance (Supplementary material 5). When the β-lactamic resistance vs β-lactamase production was compared, we found a lower percentage of β-lactamase producing strains (44% in Puebla and 45% in Sonora) than β-lactamic resistant strains. A statistical significance was found only to cefepime (p = 0.026) in strains from pregnant women in Puebla and to cefepime, aztreonam and amoxicillin-clavulanic acid (p < 0.05) in strains from pregnant and non-pregnant women from Sonora (Supplementary material 5). In the case of fluoroquinolones, there was a statistical difference in the resistance of the β-lactamase producers’ strains from pregnant women from Puebla (p ≤ 0.05) but only for norfloxacin and levofloxacin with respect to non-β-lactamase producers (Supplementary material 5).
UPEC Strains from Sonora and Puebla Can Cause Both, Lower and Upper UTIs and Their Virulence Factors Do Not Correlate with the Antibiotic Resistance
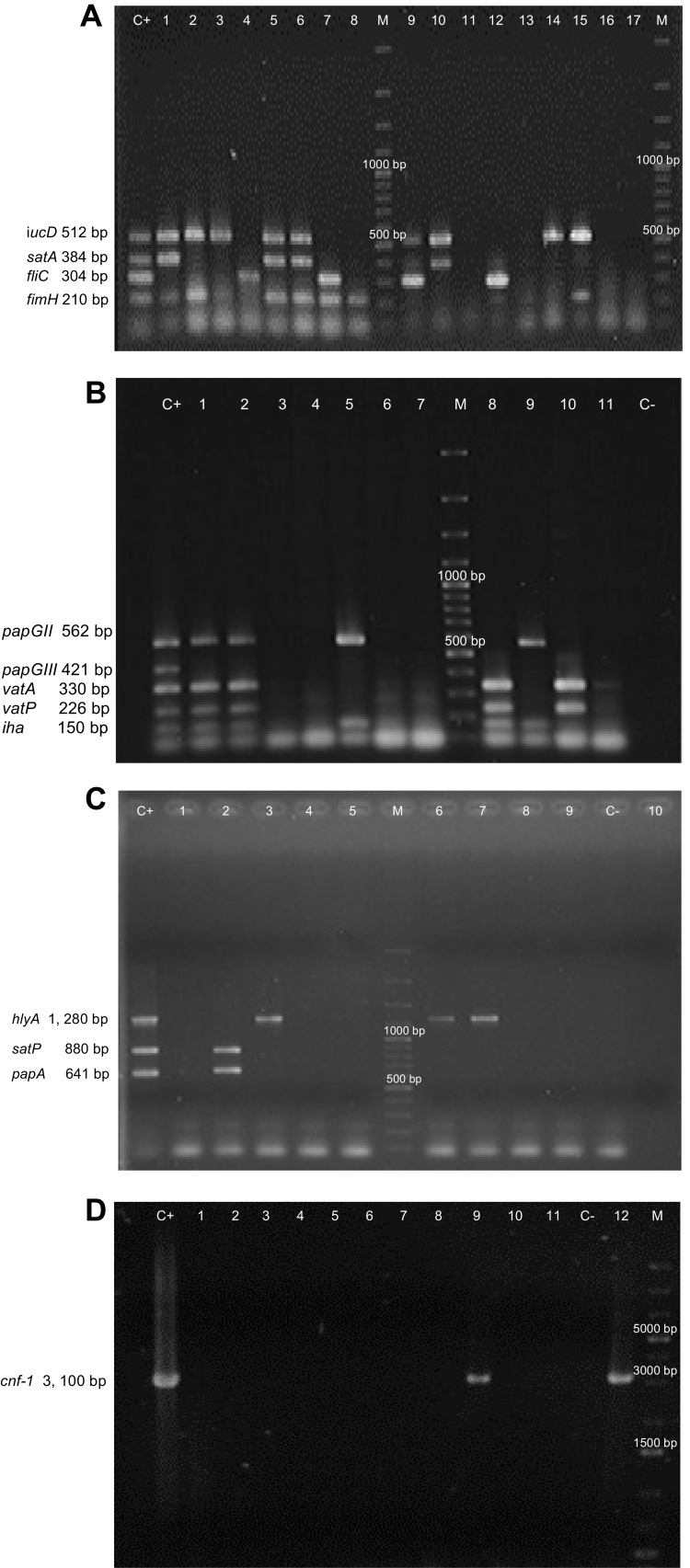
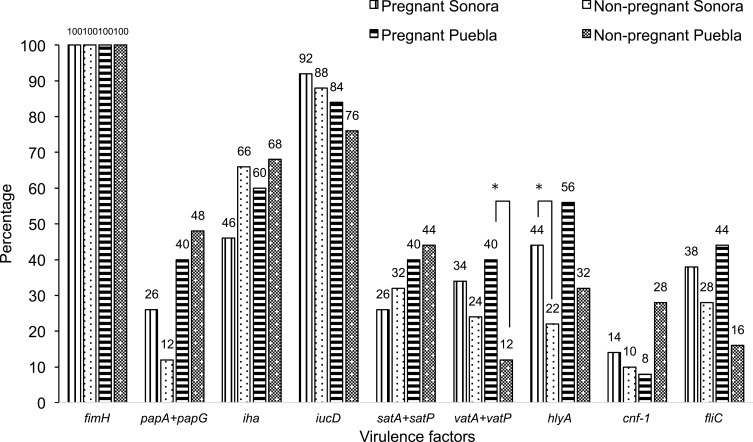
Thirteen virulence genes associated to nine virulence factors were determined by PCR (Figure 1). The most frequent virulence gene found was the type 1 pilus gene (fimH) that encodes for the pilus adhesin. This gene was found in 100% of the strains isolated from both Sonora and Puebla followed by the aerobactin receptor gene (iucD) and the iron receptor gene (iha) (Figure 2). We found the hlyA, vatA+vatP, satA+satP and fliC genes in high percentages. These genes were present in strains isolated from both states: Sonora and Puebla (Figure 2). Statistical differences between UPEC strains from pregnant vs non-pregnant women were observed only for hlyA gene (p = 0.032) in the strains isolated from Sonora and for vatA+vatP genes (p = 0.050) in the strains isolated from Puebla (Figure 2 and Supplementary material 6.1). No statistical differences were observed between the frequency and mean of virulence genes by geographic area (p > 0.05) except for papG+papA in the strains isolated from Puebla (p = 0.001) (Supplementary material 6.2). Silva et al, 2012 and Brennan et al, 2018 reported that there is an inverse relationship between antibiotic resistance and virulence.41,42 We compared the number of virulence genes with the number of antibiotics to which each strain was resistant and looked for a possible positive correlation in the strains from both Mexican states and groups, but these were only significant for strains isolated from pregnant women from Puebla (r = 0.425 and p = 0.034) (Data not shown).
Figure 1.
Multiplex-PCR Banding Patterns of Virulence Genes and Individual PCRcnf of E. coli generated by gel electrophoresis. (A) iucD (512 bp), satA (384 bp), fliC (304 bp) and fimH (210 bp). (B) papGII (562 bp), papGIII (421 bp); vatA (330 bp), vatP (226 bp) and iha (150bp). (C) hlyA (1, 280 bp), satP (880 bp) and papA (641 bp). (D) cnf-1 (3, 100 bp). M. Molecular weight market (100 bp Plus DNA Ladder in A, B and C and 1 kb Plus DNA ladder in D) . Lane C+. Positive control (E. coli CFTO73 in A; E. coli CFTO73 plus E. coli O59I in B and E. coli GAG1 in C and D) . Lines with a number E. coli isolated from the samples. C-. Negative control.
Figure 2.
Frequency of the Virulence Genes Among 150 E. coli Isolates From Urine of Pregnant and Non-Pregnant Women From Sonora and Puebla, Mexico. The studied genes encoded the following virulence factor: fimH, type 1 pilus adhesin; papG+papA, adhesin and pilin of the type P pili; iha, enterobactin receptor/Irg homologue adhesin; iucD, aerobactin; satA+satP, autotransporter and peptidase regions of the secreted autotransporter toxin; vatA+vatP, autotransporter and peptidase regions of the vacuolating autotransporter toxin; hlyA, α-hemolysin; cnf-1, cytotoxic necrotizing factor; fliC, flagellin. The statistically significance results (p < 0.05) are in asterisk.
Pathogenicity Islands (PAIs) of UPEC are Present in Strains Isolated from Sonora and Puebla
Among the mobile genetic elements responsible for the spread of virulence in UPEC strains, the PAIs stand out, which are characterized by many genes associated with virulence, so their presence indicates a high-pathogenic potential. Following the method proposed by Sabate in 2006, we searched for 4 PAIs previously reported in the UPEC prototype strains CFT073 and J96. The PAIs were found in 85 (57%) of the total of strains analyzed (n=150), 51% for Sonora´s strains (n= 100) and 68% for Puebla´s strains (n=50) (Table 3). We found these elements in 26 strains from pregnant and 25 from non-pregnant women in Sonora, while in Puebla these PAIs were found in 15 and 19 from pregnant and non-pregnant women, respectively. In both groups of strains, PAIs of E. coli CFT073 were the most prevalent (Table 3A and B), non-statistical difference was observed between distribution of these PAIs by each group and state. However, we found a higher prevalence of PAI IJ96 (33.3%) and PAI IIJ96 (53.3%) in strains from pregnant women from Puebla than from non-pregnant women in the same state without significant difference (p > 0.05) and also in strains from pregnant and non-pregnant women in Sonora (11.5% for both groups). Further, only the higher prevalence of PAI IIJ96 was statistically significant (p = 0.008) for pregnant from Puebla compared to prevalence in pregnant women from Sonora (Table 3).
Table 3.
Distribution and Percentage of Positivity of PAI in E. coli Strains Isolated from Urine of Pregnant and Non-Pregnant Women from Sonora (A) and Puebla (B), Mexico and Between States and Conditions
| PAI | A) Sonora n=51 (51%) | ||||
|---|---|---|---|---|---|
| Pregnant from Sonora n=26 | % | Non-Pregnant from Sonora n=25 | % | pb | |
| PAI ICFT073 | 19 | 73.1 | 16 | 64 | 0.77 |
| PAI IICFT073 | 17 | 65.4 | 19 | 76 | 0.54 |
| PAI IJ96 | 3 | 11.5 | 6 | 24 | 0.29 |
| PAI IIJ96 | 3 | 11.5 | 4 | 16 | 0.7 |
| PAI | B) Puebla n=34 (68%) | % | pb | ||
| Pregnant from Puebla n=15 | % | Non-pregnant from Puebla n=19 | |||
| PAI ICFT073 | 12 | 80 | 17 | 89.5 | 0.63 |
| PAI IICFT073 | 9 | 60 | 9 | 47.4 | 0.5 |
| PAI IJ96 | 5 | 33.3 | 4 | 21 | 0.46 |
| PAI IIJ96 | 8 | 53.3 | 5 | 26.3 | 0.15 |
| PAI | Pregnant from Sonora vs Pregnant from Puebla | % | pb | ||
| Pregnant from Sonora n=26 | % | Pregnant from Puebla n=15 | |||
| PAI ICFT073 | 19 | 0.77 | 12 | 80 | 0.71 |
| PAI IICFT073 | 17 | 65.4 | 9 | 60 | 0.74 |
| PAI IJ96 | 3 | 11.5 | 5 | 33.3 | 0.11 |
| PAI IIJ96 | 3 | 11.5 | 8 | 53.3 | 0.008 |
| PAI | Non-Pregnant from Sonora vs Non-Pregnant from Puebla | % | pb | ||
| Non-Pregnant from Sonora n=25 | % | Non-Pregnant from Puebla n=19 | |||
| PAI ICFT073 | 16 | 64 | 17 | 89.5 | 0.081 |
| PAI IICFT073 | 19 | 76 | 9 | 47.4 | 0.065 |
| PAI IJ96 | 6 | 24 | 4 | 21 | 1 |
| PAI IIJ96 | 4 | 16 | 5 | 26.3 | 0.467 |
Notes: pb, Fisher test exact; –, the value of p could not be obtained; In bold the statistically significant values.
Abbreviation: PAI, pathogenicity island.
Upon analyzing the frequency of PAI in all UPEC strains versus the frequencies of the genes encoding virulence factors that are localized in these PAIs such as hlyA, papG+papA and cnf-1, we found different percentages for each virulence gene. For example, cnf-1 was present in 12% of the strains from Sonora and 18% from Puebla, while the island PAIIIJ96 was found in 8% and 26%, respectively, however not statistical significance was observed between them (Supplementary material 7). This result could be because the cnf-1 gene in some strains could be present in another part of the genome, but not as part of a PAI.
UPEC Strains from Sonora and Puebla Express Functional Iron Uptakes Systems and Hemolytic Toxins
We performed phenotypic assays in order to determine siderophores and hemolysin expression using CAS agar and sheep blood agar, respectively. Iron acquisition phenotype. Our results showed that 100% of the strains isolated in both Sonora and Puebla produced some type of siderophore. We found orange halos produced by 88% of pregnant women strains and 92% of non-pregnant women strains in Puebla, while in Sonora they were observed in 74% of pregnant women strains and 94% of non-pregnant women strains. This coloration is attributed to hydroxamate type siderophores to which aerobactin (Iuc) belongs to,40 this gene (iucD) was found in 84% and 76% of the strains isolated from pregnant women and non-pregnant women in Puebla, respectively, and 92% and 88% of Sonora. We also found that 10% of strains from Puebla and 16% from Sonora presented a halo with double coloration (Yellow Orange) (Supplementary material 8.1). The slight difference in color halos between siderophore phenotype and iucD presence could be due to the production of other hydroxamate type siderophores.
Hemolysis Phenotype
In Puebla strains, the hemolytic phenotype was observed in 44% of strains from pregnant women and 28% from non-pregnant women. The phenotype was correlated with the presence of hlyA in 24% of the strains from pregnant and 8% from non-pregnant women. While in Sonora, the hemolysis phenotype was found in 38% and 56% from pregnant and non-pregnant women, respectively, with presence of hlyA in 16% and 12% respectively for each study group (Supplementary material 8.2 and Supplementary material 9).
E. coli Strains from Sonora and Puebla Belong to Serotypes Other Than UPEC and Have Variable Virulence and Resistance Properties
It is mentioned that UPEC strains belong to a limited number of serotypes. In this work, all 150 E. coli strains were serotyped. In Puebla were found 20 different serotypes (Supplementary material 10.1 and Supplementary material 10.2) corresponding to 7 pathotypes or specific clinical reports (Table 4), being predominant those related to UPEC (32%) and ETEC (10%) on average. Additionally, we found 11 (22%) no typable strains and some serotypes not reported in the literature (16%) (Table 4). The strains isolated from Sonora were grouped into 40 different serotypes (supplementary material 10.3 and Supplementary material 10.4) and 14 pathotypes or clinical reports (Table 4), these were mainly associated with UPEC (21%), STEC (10%) and EAEC (6%). Thirteen strains in this state had the O20:H9 serotype that is associated to clinical cases of neonatal sepsis. This serotype was not found in Puebla. We observed 14% and 10% of not reported serotypes and non-typable strains, respectively. These serotypes did not vary in prevalence between two study groups. In the other hand, we found a higher percentage of serotype O44:H18 (10%), associated with EAEC, in strains from non-pregnant women, followed by O6:H1 (8%), O25:H4 (8%) and O20:H9 (8%) (Supplementary material 10.4). Additionally, the serotype O25:H4 was found from Sonora´s and Puebla´s strains in 12% and 14%, respectively (Supplementary material 10.1 to 10.4). We found serotypes associated to more than one pathotype being these ones EAEC/UPEC and UPEC/STEC in both, Sonora and Puebla (Table 4). For each pathotype or clinical case associated with the serotypes found, the mean resistance to the 24 antibiotics tested and the mean virulence genes were determined (Tables 5 and 6). We found that the strains that showed resistance to a greater number of antibiotics belong to serotype associated with neonatal sepsis (O20:H9) and are from pregnant and non-pregnant women from Sonora, while pregnant and non-pregnant women from Puebla, the strains that showed resistance to a greater number of antibiotics belong to intestinal pathotypes (ETEC) and non-typable strains group (Table 5), respectively. On the other hand, the virulence gene average was 2 to 7, the strains whose serotype has been identified as STEC and indefinite pathotype were those that had a lower number of virulence genes (Table 6).
Table 4.
E. coli Strains, Isolated from Urine of Pregnant and Non-Pregnant Women from Sonora and Puebla, Associated to Pathotypes or Clinical Cases by the Serotype to Which They Belong
| Pathotype or Clinical Casea | P-SON (%) n=50 | NP-SON (%) n=50 | Accumulated n=100 (%) | P-PUE (%) n=25 | NP-PUE (%) n=25 | Accumulated n=50 (%) |
|---|---|---|---|---|---|---|
| UPEC | 12 (24) | 9 (18) | 21 (21) | 9 (36) | 7 (28) | 16 (32) |
| NT | 3 (6) | 7 (14) | 10 (10) | 7 (28) | 4 (16) | 11 (22) |
| Unreported | 5 (10) | 9 (18) | 14 (14) | 2 (8) | 6 (24) | 8 (16) |
| ETEC | – | 1 (2) | 1 (1) | 1 (4) | 4 (16) | 5 (10) |
| Heteropathogen | 2 (4) | 1 (2) | 3 (3) | 2 (8) | 1 (4) | 3 (6) |
| STEC | 4 (8) | 6 (12) | 10 (10) | 1 (4) | 1 (4) | 2 (4) |
| Isolates from diarrhea | 3 (6) | 2 (4) | 5 (5) | 2 (8) | - | 2 (4) |
| EAEC/UPEC | 3 (6) | 2 (4) | 5 (5) | - | 2 (8) | 2 (4) |
| Isolates from animals | – | 3 (6) | 3 (3) | 1 (4) | – | 1 (2) |
| EAEC | 1 (2) | 5 (10) | 6 (6) | – | – | – |
| UPEC or STEC | 4 (8) | 1 (2) | 5 (5) | – | – | – |
| Isolates from Neonatal sepsis (O20:H9) | 9 (18) | 4 (8) | 13 (13) | – | – | – |
| Isolates from healthy adults | 1 (2) | – | 1 (1) | – | – | – |
| Isolates from pyelonephritis | 1 (2) | – | 1 (1) | – | – | – |
| Isolates from SIDS | 1 (2) | – | 1 (1) | – | – | – |
| Asociado a HUS | 1 (2) | – | 1 (1) | – | – | – |
Notes: aPathotype or clinical case by the serotype to which each strain belongs. Heteropathogen, first strain of hybrid E. coli reported.
Abbreviations: UPEC, uropathogenic E. coli; NT, Notypable; STEC, Shiga toxigenic E. coli; EAEC, enteroaggregative E. coli; EPEC, enteropathogenic E. coli; ETEC, Enterotoxigenic E. coli; SIDS, sudden infant death syndrome; HUS, hemolytic uremic syndrome; P-SON, pregnant women from Sonora; NP-SON, Non-pregnant women from Sonora; P-PUE, pregnant women from Puebla; NP-PUE, non-pregnant women from Puebla.
Table 5.
Antibiotic Resistance of E. coli Strains Associated to Pathotypes or Clinical Cases by the Serotype to Which They Belong
| Pathotype or Clinical Casea | Mean of Antibiotic Resistanceb | |||
|---|---|---|---|---|
| P-SON |
NP-SON |
P-PUE |
NP-P |
|
| UPEC | 15 | 15 | 13 | 15 |
| NT | 13 | 12 | 15 | 16 |
| Unreported | 14 | 14 | 15 | 16 |
| ETEC | – | 10 | 19 | 10 |
| Heteropathogen | 12 | 14 | 15 | 13 |
| STEC | – | 13 | 12 | 11 |
| Isolates from diarrhea | 16 | 12 | 18 | – |
| EAEC, EPEC | 11 | 12 | – | 16 |
| UPEC, STEC | 12 | – | – | – |
| Isolates from animals | – | 14 | 17 | – |
| EAEC | 11 | 14 | – | – |
| Isolates from neonatal sepsis (O20:H9) | 20 | 19 | – | - |
Notes: aPathotype or clinical case by the serotype to which each strain belongs. Heteropathogen, first strain of hybrid E. coli reported; the higher mean of antibiotic resistance are in bold. b24 groups of antibiotics tested.
Abbreviations: STEC, shiga toxin productor E. coli; UPEC, uropathogenic E. coli; NT, Notypable; ETEC, Enterotoxigenic E. coli; EAEC, Enteroaggregative E. coli; EPEC, Enteropathogenic E. coli; b, 24 groups of antibiotics tested; P-SON, pregnant women from Sonora; NP-SON, Non-pregnant women from Sonora; P-PUE, pregnant women from Puebla; NP-PUE, Non-pregnant women from Puebla.
Table 6.
Virulence Genes of E. coli Strains Associated to Pathotypes or Specific Clinic Cases by the Serotype to Which They Belong
| Pathotype or Clinic Casea | Mean of Virulence Genesb | |||
|---|---|---|---|---|
| P-SON | NP-SON | P-PUE | NP-PUE | |
| UPEC | 5 | 5 | 6 | 3 |
| NT | 4 | 3 | 5 | 4 |
| Unreported | 4 | 4 | 4 | 5 |
| ETEC | – | 4 | 4 | 6 |
| Heteropathogen (O2:H6) | 5 | 7 | 4 | 7 |
| STEC | 2 | 4 | 6 | 2 |
| Isolates from diarrhea | 5 | 4 | 4 | – |
| Indefinite (EAEC, EPEC, UPEC) (O15:H18) | 4 | 3 | – | 4 |
| UPEC, STEC | 5 | – | – | – |
| EAEC | 4 | 3 | – | – |
| Isolates from neonatal sepsis (O20:H9) | 3 | 4 | – | – |
| Others | 5 | 5 | 6 | – |
Notes: aPathotype or clinical case by the serotype to which each strain belongs. bNine virulence factors tested by PCR; Others, Included Isolates from animals, isolates from healthy adults, isolates from hemolytic uremic syndrome and isolates from sudden infant death syndrome.
Abbreviations: UPEC, Uropathogenic E. coli; ETEC, Enterotoxigenic E. coli; STEC, Shiga toxigenic E. coli; EAEC, enteroaggregative E. coli; EPEC, enteropathogenic E. coli; NT, notypable; P-SON, pregnant women from Sonora; NP-SON, Non-pregnant women from Sonora; P-PUE, pregnant women from Puebla; NP-PUE, Non-pregnant women from Puebla.
Virulence and Antibiotic Resistance are Not Related to Specifics Phylogroups
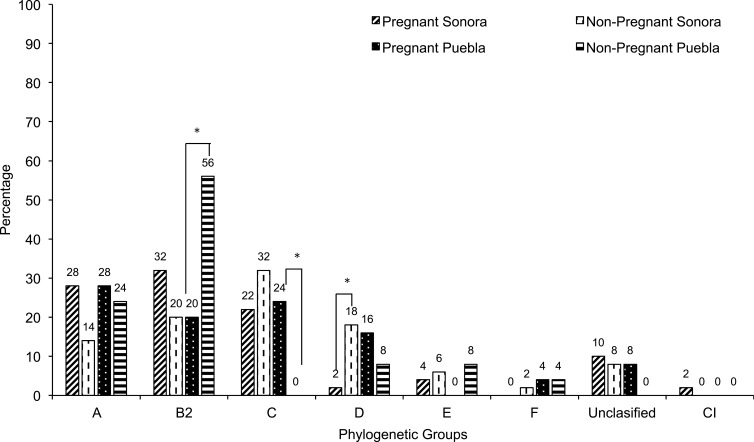
Differences were found in the distribution of the phylogenetic groups. In Sonora, the results obtained from pregnant women showed a higher prevalence of strains belonging to the phylogroups B2 (32%), A (28%) and C (22%). In this group, we observed one strain that was classified within clade I that are strains belonging to Escherichia, but not coli, in addition 10% could not be assigned to any phylogenetic group (Figure 3). While in non-pregnant women the phylogenetic group C was found in 32%, followed by B2 (20%), D (18%), A (14%), E (6%) and F (2%). A small percentage of strains (8%) could not be assigned phylogenetically to any group. On the other hand, in pregnant women from Puebla, we found a higher percentage of strains belonging to phylogroup A (28%), followed by C (24%) and B2 (20%), phylogroups D and F were found in 16% and 4%, respectively. While the strains isolated from non-pregnant women belonged mainly to phylogroups B2 (56%) and A (24%) (Figure 3). Additionally, we found within of this same samples 8% (n=2) of strains that were not possible to group phylogenetically. Statistically significance was found in the highest percentage of the phylogroups B2 (p = 0.018) in strains from non-pregnant women in Puebla, C (p = 0.022) in strains from pregnant women in Puebla and D (p = 0.015) in strains from non-pregnant women in Sonora (Figure 3). No statistically significant differences were found in the distribution of phylogroups by geographical area (p > 0.05), except for group D, whose frequency was higher in the isolates of pregnant women in Puebla with respect to those in Sonora (p = 0.039). This also happened with the phylogroups B2 in the Sonora strains (p = 0.003) and C in Puebla (<0.001), in both cases isolated from pregnant patients (Data not shown).
Figure 3.
Distribution of phylogenetic group among 150 E. coli strains isolated from urine of women from Sonora and Puebla, Mexico. The statistically significance results (p<0.05) are in asterisk.
Abbreviation: CI, Clade I.
When comparing the mean of virulence genes of the strains according to the phylogenetic groups and the mean number of antibiotics to which they resist, no statistically significant differences were found (Data not shown).
Discussion
The growing bacterial antibiotic resistance in recent years implies a serious health problem that has reduced the options for infection treatments, including UTIs. We found that 92.7% of the strains from pregnant and non-pregnant women isolated in both states were classified as multidrug-resistant, and a small percentage was classified as XDR and PDR. Our results regarding MDR and XDR were higher than those reported by Ochoa in 2016 in Mexico, in which only 10 antibiotics were used43 and the PDR strains were not found. The susceptibility profiles indicated that strains isolated from women in both states have a high resistance to β-lactam antibiotics, aminoglycosides, nitrofurans and amoxicillin-clavulanic acid, these results were similar to those reported in Mexico (Guerrero and Mexico city) and in Nigeria.12,44 It becomes of relevant importance for pregnant woman since these antibiotics are in the short list of antibiotics available for the treatment of lower and upper UTI in this group. Our results showed a considerable reduction in the number of antibiotics available for UTIs treatment, showing the necessity of searching for other therapeutic alternatives as well as continuous monitoring of the empirical treatments implemented in Mexico. Alternatively, implementation of individualized treatments according to the results obtained in the antibiotic susceptibility profile, could avoid the emergence of more resistant and multi-resistant UPEC strains or prevent the co-selection phenomenon.
One of the resistance mechanisms for broad-spectrum penicillins, cephalosporins and monobactams widely disseminated in UPEC is the production of β-lactamases. We found ESBL production in 68% of strains isolated from pregnant women and 20% from non-pregnant women in Puebla, while in Sonora it was found in 44% of strains from pregnant women and 46% from non-pregnant women. The production of these enzymes in strains from pregnant patients has been reported by other researchers such as Ramos in 2012 and Al-Mayahie in 2013, but these authors report the phenotype in lower percentage than what we documented.45,46 Interestingly, in the present study, the percentage of ESBL phenotype was lower than the percentage of strains resistant to β-lactams, this could indicate that UPEC strains could present other mechanisms of resistance to this family of antibiotics, as the production of an altered penicillin binding proteins (PBP) which decrease the affinity of the antibiotic for PBP or efflux pumps that lead to bacterial survival and therapeutic failure.47,48
The predominant virulence factors present in pregnant and non-pregnant women of both states were those involved in adhesion and iron acquisition. All strains isolated from both Sonora and Puebla presented the fimbrial adhesin of type 1 pili (fimH), which has been associated with events not only of adherence to the bladder but also linked to internalization and subsequent formation of intracellular bacterial communities (IBC), which are associated with recurrent UTI episodes, antibiotic resistance and immunoevasion.49–51 Furthermore, a remarkable predominance of other important virulence genes, such as fliC (44%), papG+papA (40%), hlyA (56%), satA+satP (40%) and vatA+vatP (40%) can be seen in isolated strains from pregnant women in Puebla. The products of these genes are involved in upper UTI, indicating the high potential of pathogenicity of these strains. Our results from both Sonora and Puebla resemble those obtained by Miranda-Estrada in Mexico city and Guerrero while the genes involved in adherence were found in ranges of 25% −100% in Mexico city and 10% −78% in the Guerrero state.12 However, our study found a higher prevalence of these genes than the Miranda-Estrada study. It is worrisome to observe virulence features associated with upper UTI in strains isolated from both groups of patients, but mainly in pregnant women. In this sense, we cannot assure that the strains are causing upper urinary infections in these patients. Although hlyA and fliC are determinants that have been observed in pyelonephritic clinical isolates and a positive phenotype was observed in several isolates, the phenotypic test for the expression of the P-type pili is still pending, together with the fact that we do not have the clinical history of the patients prevents us from concluding that they are pyelonephritic strains, but we can conclude that they have a high potential (given their genetic load, hemolysis phenotype, mobility and the group of patients from which they were isolated) to cause upper urinary tract infections. Additionally, it is important to mention that given changes such as increased glomerular filtration, trigone hypertrophy and subsequent bladder-ureteral reflux in conjunction with the virulence load of the strains increase the likelihood that they may be causing upper UTI in pregnant patients.52,53 So, it would be interesting in future studies to know the clinical characteristics of these patients to associate them with the virulence phenotypes and genotypes of isolated UPEC strains causing UTI. Additionally, some authors mentioned that there is an inverse relationship between antibiotic resistance and virulence.41,42 However, we found the opposite in strains isolated from pregnant women in Puebla where there was a positive correlation between virulence and resistance. With this observation, we can conclude that the virulence and resistant of E. coli strains from Mexican women are not always associated.
We found a high percentage of positive phenotypes for iron acquisition (92% in Puebla and 88% in Sonora), this can be explained by the importance of iron for the survival of UPEC in the urinary tract, while the slight difference between siderophore phenotype and iucD presence could be due to the production of other hydroxamate siderophores such as ferrichrome, coprogen, and rhodotorulic acid that have been reported in E. coli strains.54,55 In the other hand, the low percentage of coincidence between phenotype and genotype of hemolysis could be due to an incomplete process of activation of the pro-toxin (HlyA), which does not allow the formation of the corresponding heptameter in the target cell and therefore the phenotype could not be appreciated.56,57
Among the mobile-genetic elements responsible for the spread of virulence in UPEC strains, PAIs are important players with high pathogenic potential.36 We found a higher prevalence of CFT073 PAIs compared to J96 PAIs. This result coincides with the reported by other authors in Spain, Iran and Iraq.58–60 However, in our work, in addition to high percentage of CFT073 islands, we found, a higher percentage of PAIs (I and II) from E. coli J96 in both states, than the authors above mentioned. These results suggest that some E. coli strains isolated from Mexican patients with UTIs have PAIs more associated to E. coli J96 than the strains isolated in the mentioned countries, although more studies need to be done in Mexico to reinforce our hypothesis.
UPEC strains have been associated to a small number of serotypes;9 however, we found a great variety, including some associated with other pathotypes, which make us propose that in Mexico, UPEC is not so restricted. We found serotypes associated with intestinal strains, but the virulence factors present are related to the urinary infection processes. We do not rule out the possibility that some of these strains have also factors associated with intestinal strains, because the existence of hybrid strains have been reported and their virulence characteristics corresponded to those of intestinal and extra-intestinal strains. Among the serotypes observed, we found O2:H6 associated with the first reported hybrid E. coli strain (STEC/UPEC).63 Therefore, it would be interesting to find out whether the genes of diarreaghenic E. coli are present in ours strains that are associated with these serotypes rather than those reported as UPEC. We also found a considerable percentage (22% Puebla and 10% Sonora) of non-typable strains, which could be suggestive of new serotypes present in Mexico. Interestingly, some serotypes that were found have been associated with disease conditions in animals and unconventional situations such as corpses of infants with sudden death syndrome64,65. With these results, we could propose that UPEC strains are not serotypically restricted, because despite having found a great variety of serotypes, some are non-typables or are associated with other pathotypes.
It has been previously reported that the predominant phylogenetic groups in pathogenic strains for humans are B2 and D, while A corresponds to commensal strains which are a reservoir of the resistance genes.37,66 Previous research conducted in Mexico, Nigeria, Uganda, Sweden, Vietnam, India and Mongolia,12,16,44,45 have reported the prevalence of the phylogenetic group B2 (ranges from 33.8% −58%), followed by A (5% −28%) and D (5% −28%). When analyzing these studies, we noticed a variation in the prevalence of these phylogenetic groups according to the geographical area. In our work, we observed different characteristics to those reported by these authors; in pregnant women from Puebla, we found a higher percentage of strains belonging to the phylogenetic group A (28%), while in Sonora, the phylogenetic groups B2 (32%) and A (28%) were observed. In strains obtained from non-pregnant women, phylogroup B2 (56%) was predominant in Puebla, while in Sonora it was group C (32%). Interestingly, when analyzing the means of virulence and resistance, we found that the phylogroup C has a comparable degree of virulence and resistance and even superior to that presented by strains of phylogroups considered pathogenic for humans. Our results are important because it is one of the few studies that use the new phylogenetic classification scheme reported by Clermont 2012. However, is evident that group A, it is not only a reservoir of antibiotic resistance, but it harbors specific virulence characteristics to cause UTI.
Conclusion
This is the first study conducted in Mexico in which virulence, resistance, phylogenetic groups and serotypes of UPEC strains isolated from pregnant and non-pregnant women are compared in two-Mexican states. UPEC strains from women with UTIs in Puebla and Sonora were multidrug resistant, including to antibiotics of first choice in treatment, with a tendency to become XDR or PDR. The resistance profiles were varied by geographical area, demonstrating the importance of change from empirical to individualized treatment and the enormous need to search for new therapeutic alternatives for the treatment of UTIs. E. coli strains were serotypically and phylogenetically variable and there are a good percentage of unknown serotypes and phylogroups with virulence characteristics of UPEC, which could imply new serotypes and other phylogroups associated with this important pathogen. The results also showed that UPEC strains isolated from pregnant and no-pregnant women have virulence factors that are characteristic or presumptively limited to UPEC strains that cause lower UTIs. Nevertheless, a considerable portion of strains mainly from pregnant women had optimal virulence characteristics to cause upper UTIs. These results will allow us to better understand the behavior of UPEC as an etiological agent of UTIs in the female population in Mexico and for better treatment and control of the disease and emphasize the need of future research in Mexico.
Acknowledgments
We thank to PhD Alfredo G. Torres Tejeda (University of Texas, Medical Branch (UTMB)) for the review of the manuscript and his important comments and contribution. Thanks to Ms. N´machi Anumba from Vicerrectoria de Investigación y Estudios de posgrado (VIEP), BUAP for proofreading the manuscript. We would like to thank DC Lucia Soto Urzua, MC José Angel F. Flores-Hernández and DC Lorena Luna Guevara (BUAP) for technical assistance. We would like to thank Paul Gaytán, Jorge Yañez, Eugenio Bustos and Santiago Becerra (Unidad de Síntesis y Secuenciación IBT-UNAM) for DNA oligonucleotides syntheses used in this study. The laboratory of M.M.P.A.-H and R. C. R. G. were supported in part by institutional funds from the Programa Institucional para la Consolidación de los Cuerpos Académicos y Conformación de Redes de Investigación 100182644-VIEP2019 and from Centro de Investigación en Ciencias Microbiológicas del Instituto de Ciencias, BUAP (CICM-ICUAP and Postgrado en Microbiología (CICM-ICUAP). The author M. G. B. M. had CONACYT Master Fellowship during the performance of this work (Scholarship No. 617232).
Disclosure
The authors declare that they have no conflict interests in this work.
References
- 1.Dirección General de Epidemiología. Boletín Epidemiológico. Sistema Nacional De Vigilancia Epidemiológica. Vol. 34 México, D.F: Sistema Único de Información; 2017. [Google Scholar]
- 2.Hill JB, Sheffield JS, Mcintire DD. Acute pyelonephritis in pregnancy. Am Fam Physician. 2005;42(4):1069. doi: 10.1097/01.AOG.0000149154.96285.a0 [DOI] [PubMed] [Google Scholar]
- 3.Andreu A, Cacho J, Coira A, Lepe JA. Diagnóstico microbiológico de las infecciones del tracto urinario. Enferm Infecc Microbiol Clin. 2011;29(1):52–57. doi: 10.1016/j.eimc.2010.06.008 [DOI] [PubMed] [Google Scholar]
- 4.Gilbert NM, O’brien VP, Hultgren S, Macones G, Lewis WG, Lewis AL. Urinary tract infection as a preventable cause of pregnancy complications: opportunities, challenges, and a global call to action. Glob Adv Heal Med. 2013;2(5):59–69. doi: 10.7453/gahmj.2013.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielubowicz GR, Mobley HLT. Host–pathogen interactions in urinary tract infection. Nat Rev Urol. 2010;7(8):430–441. doi: 10.1038/nrurol.2010.101 [DOI] [PubMed] [Google Scholar]
- 6.Hannan TJ, Totsika M, Mansfield KJ, Moore KH, Mark A, Hultgren SJ. Host-pathogen checkpoints and population bottlenecks in persistent and intracellular uropathogenic E. coli bladder infection. NIH Public Access. 2013;36(3):616–648. doi: 10.1111/j.1574-6976.2012.00339.x.Host-Pathogen [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerure RD. Prevalence of asymptomatic bacteriuria among pregnant women in a tertiary care hospital. J Clin Diagn Res. 2013;3(11):11–14. [Google Scholar]
- 8.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13(5):269–284. doi: 10.1038/nrmicro3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiles TJ, Kulesus RR, Mulvey MA. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp Mol Pathol. 2008;85:11–19. doi: 10.1016/j.yexmp.2008.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sainz E, Rosario T, Reyes M, Vicente P, Patricia M, Eslava C. Resistencia a antimicrobianos de cepas de E. coli de diversos serotipos aisladas de pacientes de un Hospital Psiquiátrico. Rev Mex Ciencias Farm. 2008;39(4):18–25. [Google Scholar]
- 11.Millán Y, Hernández E, Millán B, Araque M. Distribución de grupos filogenéticos y factores de virulencia en cepas de Escherichia coli uropatógena productora de β-lactamasa CTX-M-15 aisladas de pacientes de la comunidad en Mérida, Venezuela. Rev Argent Microbiol. 2014;46(3):175–181. doi: 10.1016/S0325-7541(14)70069-0 [DOI] [PubMed] [Google Scholar]
- 12.Miranda-Estrada LI, Ruíz-Rosas M, Molina-López J, Parra-Rojas I, González-Villalobos E, Castro-Alarcón N. Relación entre factores de virulencia, resistencia a antibióticos y los grupos filogenéticos de Escherichia coli uropatógena en dos localidades de México. Enferm Infecc Microbiol Clin. 2017;35(7):426–433. doi: 10.1016/j.eimc.2016.02.021 [DOI] [PubMed] [Google Scholar]
- 13.Hojati Z, Molaie R, Gholipour A. The FimH gene in uropathogenic Escherichia coli strains isolated from patient with urinary tract infection. Jundishapur J Microbiol. 2015;8(2). doi: 10.5812/jjm.17520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahdar M, Rashki A, Miri HR, Ghalehnoo MR. Detection of pap, sfa, afa, foc, and fim adhesin-encoding operons in uropathogenic Escherichia coli isolates collected from patients with urinary tract infection. Jundishapur J Microbiol. 2015;8(8). doi: 10.5812/jjm.22647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Safi M, Achour W, Baaboura R, El Fatmi R, Ben Othmen T, Ben Hassen A. Distribution of virulence associated traits among urine Escherichia coli isolates from patients in onco-hematology. J Infect Chemother. 2016;22(4):221–224. doi: 10.1016/j.jiac.2015.12.017 [DOI] [PubMed] [Google Scholar]
- 16.Munkhdelger Y, Gunregjav N, Dorjpurev A, Juniichiro N, Sarantuya J. Detection of virulence genes, phylogenetic group and antibiotic resistance of uropathogenic Escherichia coli in Mongolia. J Infect Dev Ctries. 2017;11(01):51. doi: 10.3855/jidc.7903 [DOI] [PubMed] [Google Scholar]
- 17.Rahdar M, Rashki A, Miri HR, Rashki Ghalehnoo M. Detection of pap, sfa, afa, foc, and fim adhesin-encoding operons in uropathogenic Escherichia coli isolates collected from patients with urinary tract infection. Jundishapur J Microbiol. 2015;8(8). doi: 10.5812/jjm.22647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ali I, Rafaque Z, Ahmed S, Malik S, Dasti JI. Prevalence of multi-drug resistant uropathogenic Escherichia coli in Potohar region of Pakistan. Asian Pac J Trop Biomed. 2016;6(1):60–66. doi: 10.1016/j.apjtb.2015.09.022 [DOI] [Google Scholar]
- 19.Bien J, Sokolova O, Bozko P. Role of uropathogenic Escherichia coli virulence factors in development of urinary tract infection and kidney damage. Int J Nephrol. 2012. doi: 10.1155/2012/681473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Figueroa-Damián R, Velásquez P, Carrera A, Hernández-Valencia M. Consenso para el tratamiento de infecciones urinarias en ginecología. Perinat Reprod Hum. 2008;22:22–39. [Google Scholar]
- 21.Estrada AA, Figueroa DR, Villagana ZR. Infección de vías urinarias en la mujer embarazada. Importancia del escrutinio de bacteriuria asintomática durante la gestación. Perinatol Reprod Humana. 2010;24(3):182–186. [Google Scholar]
- 22.Calderón-Jaimes E. Diagnóstico y tratamiento de las infecciones en vías urinarias: un enfoque multidisciplinario para casos no complicados. Bol Med Hosp Infant Mex. 2013;70(1):3–10. [Google Scholar]
- 23.Salud S. Guía de Práctica Clínica: prevención, Dianóstico y Tratamiento de La Infección Del Tracto Urinario Bajo, Durante El Embarazo, En El Primer Nivel de Atencion [Clinical Practice Guide: Prevention, Diagnosis and Treatment of Low Urinary Tract Infection, During Pregnancy, At the First Level of Care]. 2016. Available from: http://www.cenetec-difusion.com/CMGPC/IMSS-078-08/ER.pdf. Accessed December31, 2019.
- 24.Vallano A, Arnau JM. Antimicrobianos y embarazo. Enferm Infecc Microbiol Clin. 2009;27(9):536–542. doi: 10.1016/j.eimc.2009.09.001 [DOI] [PubMed] [Google Scholar]
- 25.World Health Organization. Antibiotic resistance. Available from: http://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance. Accessed December31, 2019 Published 2018.
- 26.Alqasim A, Abu Jaffal A, Alyousef AA. Prevalence of multidrug resistance and extended-spectrum β -lactamase carriage of clinical uropathogenic Escherichia coli isolates in Riyadh, Saudi Arabia. Int J Microbiol. 2018;2018:1–9. doi: 10.1155/2018/3026851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malekzadegan Y, Khashei R, Sedigh Ebrahim-Saraie H, Jahanabadi Z. Distribution of virulence genes and their association with antimicrobial resistance among uropathogenic Escherichia coli isolates from Iranian patients. BMC Infect Dis. 2018;18(1):1–9. doi: 10.1186/s12879-018-3467-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bajpai T, Pandey M, Varma M, Bhatambare G. Prevalence of TEM, SHV, and CTX-M Beta-Lactamase genes in the urinary isolates of a tertiary care hospital. Avicenna J Med. 2017;7(1):12. doi: 10.4103/2231-0770.197508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bush K, Jacoby GA. Updated functional classification of β-lactamases. Antimicrob Agents Chemother. 2010;54(3):969–976. doi: 10.1128/AAC.01009-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonomo RA. β-lactamases: A Focus on Current Challenges. Cold Spring Harb Perspect Med. 2017;7:a025239. doi: 10.1101/cshperspect.a025239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Organizacion Mundial de la Salud. Guía sobre la reglamentación relativa al transporte de sustancias infecciosas 2013–2014: aplicable a partir de 1 de enero de 2013. Plant Dis. 2012;96. doi: 10.1094/PDIS-11-11-0999-PDN [DOI] [Google Scholar]
- 32.Ørskov F, Ørskov I. 2 Serotyping of Escherichia coli. Methods Microbiol. 1984;43–112. doi: 10.1016/S0580-9517(08)70447-1 [DOI] [Google Scholar]
- 33.Sambrook J. Molecular Cloning a Laboratory Manual. Vol. 33 2012. doi: 10.3724/SP.J.1141.2012.01075 [DOI] [Google Scholar]
- 34.CLSI. Performance Standards for Antimicrobial Susceptibility Testing. Twenty-Fifth Informational Supplement; 2017. [Google Scholar]
- 35.Magiorakos A, Srinivasan A, Carey RB, et al. Bacteria: an international expert proposal for interim standard definitions for acquired resistance. Microbiology. 2011;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 36.Sabate M, Moreno E, Perez T, Andreu A, Prats G. Pathogenicity island markers in commensal and uropathogenic Escherichia coli isolates. Clin Microbiol Infect. 2006;12(9):880–886. doi: 10.1111/j.1469-0691.2006.01461.x [DOI] [PubMed] [Google Scholar]
- 37.Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. 2013;5(1):58–65. doi: 10.1111/1758-2229.12019 [DOI] [PubMed] [Google Scholar]
- 38.Milagres AMF, Machuca A, Napoleão D. Detection of siderophore production from several fungi and bacteria by a modification of chrome azurol S (CAS) agar plate assay. J Microbiol Methods. 1999;37(1):1–6. doi: 10.1016/S0167-7012(99)00028-7 [DOI] [PubMed] [Google Scholar]
- 39.Shin SH, Lim Y, Lee SE, Yang NW, Rhee JH. CAS agar diffusion assay for the measurement of siderophores in biological fluids. J Microbiol Methods. 2001;44(1):89–95. doi: 10.1016/S0167-7012(00)00229-3 [DOI] [PubMed] [Google Scholar]
- 40.Pérez-Miranda S, Cabirol N, George-Téllez R, Zamudio-Rivera LS, Fernández FJ. O-CAS, a fast and universal method for siderophore detection. J Microbiol Methods. 2007;70(1):127–131. doi: 10.1016/j.mimet.2007.03.023 [DOI] [PubMed] [Google Scholar]
- 41.da Silva GJ, Mendonça N. Association between antimicrobial resistance and virulence in Escherichia coli. Virulence. 2012;3(1):18–28. doi: 10.4161/viru.3.1.18382 [DOI] [PubMed] [Google Scholar]
- 42.Brennan DE, Dowd C, O’Morain C, McNamara D, Smith SM. Can bacterial virulence factors predict antibiotic resistant Helicobacter pylori infection? World J Gastroenterol. 2018;24(9):971–981. doi: 10.3748/wjg.v24.i9.971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ochoa SA, Cruz-Córdova A, Luna-Pineda VM, et al. Multidrug- and extensively drug-resistant uropathogenic Escherichia coli clinical strains: phylogenetic groups widely associated with integrons maintain high genetic diversity. Front Microbiol. 2016;7(DEC). doi: 10.3389/fmicb.2016.02042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abiodun AO, Olufunke OA, Dunah FC, Oladiran F. Phenotypic identification and phylogenetic characterization of uropathogenic escherichia coli in symptomatic pregnant women with urinary tract infections in South-Western Nigeria. Int J Biol. 2014;6(4). doi: 10.5539/ijb.v6n4p145 [DOI] [Google Scholar]
- 45.Ramos NL, Sekikubo M, Dzung DTN, et al. Uropathogenic Escherichia coli isolates from pregnant women in different countries. J Clin Microbiol. 2012;50(11):3569–3574. doi: 10.1128/JCM.01647-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Al-Mayahie SMG. Phenotypic and genotypic comparison of ESBL production by vaginal Escherichia coli isolates from pregnant and non-pregnant women. Ann Clin Microbiol Antimicrob. 2013;12:7. doi: 10.1186/1476-0711-12-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rice LB. Mechanisms of resistance and clinical relevance of resistance to β-lactams, glycopeptides, and fluoroquinolones. Mayo Clin Proc. 2012;87(2):198–208. doi: 10.1016/j.mayocp.2011.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Worthington RJ, Melander C. Overcoming resistance to β-lactam antibiotics. J Org Chem. 2013;78(9):4207–4213. doi: 10.1021/jo400236f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eto DS, Jones TA, Sundsbak JL, Mulvey MA. Integrin-mediated host cell invasion by type 1–piliated uropathogenic Escherichia coli. PLoS Pathog. 2007;3(7):e100. doi: 10.1371/journal.ppat.0030100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinez JJ, Hultgren SJ. Requirement of Rho-family GTPases in the invasion of type 1-piliated uropathogenic Escherichia coli. Cell Microbiol. 2002;4(1):19–28. doi: 10.1046/j.1462-5822.2002.00166.x [DOI] [PubMed] [Google Scholar]
- 51.Guiton PS, Cusumano CK, Kline KA, et al. Combinatorial small-molecule therapy prevents uropathogenic Escherichia coli catheter-associated urinary tract infections in mice. Antimicrob Agents Chemother. 2012;56(9):4738–4745. doi: 10.1128/AAC.00447-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Autún Rosado DP, Sanabria Padrón VH, Figueroa EHC, Rangel Villaseñor O, Hernández-Valencia M. Etiología y frecuencia de bacteriuria asintomática en mujeres embarazadas. Perinatol Reprod Hum. 2015;29(4):148–151. doi: 10.1016/j.rprh.2016.02.001 [DOI] [Google Scholar]
- 53.Vallejos Medic C, López Villegas MDR, Enríquez Guerra MÁ, Ramírez Valverde B. Prevalencia de infecciones de vías urinarias en embarazadas atendidas en el Hospital Universitario de Puebla. Enfermedades Infec Microbiol. 2010;30(4):118–122. [Google Scholar]
- 54.Hantke K. Identification of an iron uptake system specific for coprogen and rhodotorulic acid in Escherichia coli K12. Mol Gen Genet. 1983;191:301–306. doi: 10.1007/BF00334830 [DOI] [PubMed] [Google Scholar]
- 55.Garcia EC, Brumbaugh AR, Mobley HLT. Redundancy and specificity of escherichia coli iron acquisition systems during urinary tract infection. Payne SM, ed. Infect Immun. 2011;79(3):1225–1235. doi: 10.1128/IAI.01222-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Herlax V, Maté S, Rimoldi O, Bakás L. Relevance of fatty acid covalently bound to escherichia coli α-hemolysin and membrane microdomains in the oligomerization process. J Biol Chem. 2009;284(37):25199–25210. doi: 10.1074/jbc.M109.009365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bakás L, Maté S, Vazquez RHV. E. coli alpha-hemolysin and properties In: Ekinci D, editor. Biochemistry. Rijeka, Croacia: InTech; 2012:107–140. [Google Scholar]
- 58.Naderi G, Haghi F, Zeighami H, Hemati F, Masoumian N. Distribution of pathogenicity island (PAI) markers and phylogenetic groups in diarrheagenic and commensal Escherichia coli from young children. Gastroenterol Hepatol Bed Bench. 2016;9(4):316–324. [PMC free article] [PubMed] [Google Scholar]
- 59.Messerer M, Fischer W, Schubert S. Investigation of horizontal gene transfer of pathogenicity islands in Escherichia coli using next-generation sequencing. PLoS One. 2017;12(7):1–17. doi: 10.1371/journal.pone.0179880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Navidinia M, Najar Peerayeh S, Fallah F, et al. Distribution of the pathogenicity islands markers (PAIs) in uropathogenic E.coli isolated from children in mofid children hospital. Arch Pediatr Infect Dis. 2013;1(2):75–79. doi: 10.5812/pedinfect.9083 [DOI] [Google Scholar]
- 61.Samei A, Haghi F, Zeighami H. Distribution of pathogenicity island markers in commensal and uropathogenic Escherichia coli isolates. Folia Microbiol (Praha). 2016;61(3):261–268. doi: 10.1007/s12223-015-0433-8 [DOI] [PubMed] [Google Scholar]
- 62.Tangi SC, Tajbakhsh E, Soleimani NA, Shahraki MM. Prevalence of pathogenicity island markers genes in uropathogenic Escherichia coli isolated from patients with urinary tract infectious. Asian Pacific J Trop Dis. 2015;5(8):662–666. doi: 10.1016/S2222-1808(15)60909-4 [DOI] [Google Scholar]
- 63.Bielaszewska M, Schiller R, Lammers L, et al. Heteropathogenic virulence and phylogeny reveal phased pathogenic metamorphosis in Escherichia coli O2: H6. EMBO Mol Med. 2014;6(3):347–357. doi: 10.1002/emmm.201303133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Poppe C, Martin L, Gyles C, et al. Acquisition and transfer of resistance to extended-spectrum cephalosporins by Salmonella Newport and E. coli in the intestinal tract of turkey poults. Guelph Food Saf Semin Ser Symp Guelph. 2004;71(3):1184–1192. doi: 10.1128/AEM.71.3.1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pearce JL, Bettelheim KA, Luke RKJ, Goldwater PN. Serotypes of Escherichia coli in sudden infant death syndrome. J Appl Microbiol. 2010;108(2):731–735. doi: 10.1111/j.1365-2672.2009.04473.x [DOI] [PubMed] [Google Scholar]
- 66.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66(10):4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Salud S. Guía de Práctica Clínica: prevención, Dianóstico y Tratamiento de La Infección Del Tracto Urinario Bajo, Durante El Embarazo, En El Primer Nivel de Atencion [Clinical Practice Guide: Prevention, Diagnosis and Treatment of Low Urinary Tract Infection, During Pregnancy, At the First Level of Care]. 2016. Available from: http://www.cenetec-difusion.com/CMGPC/IMSS-078-08/ER.pdf. Accessed December31, 2019.