Figure 4.
FoxO1 and miR-183-5p Are Downstream Targets of mmu_circ_0001359
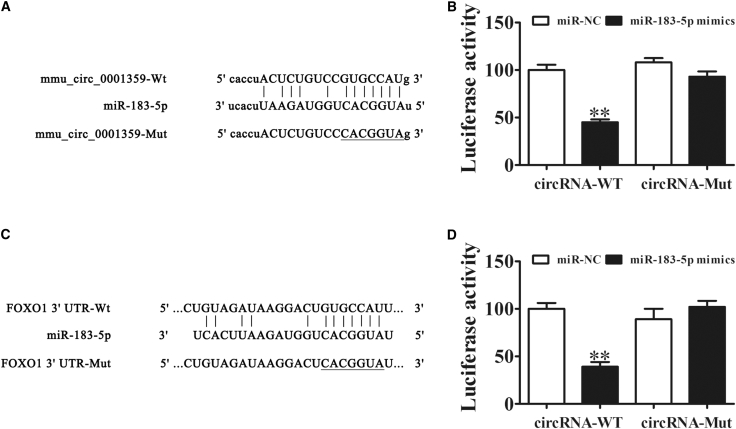
(A) Complementary sequences in miR-183-5p and mmu_circ_0001359 were obtained using publicly available algorithms. A mutated version of mmu_circ_0001359 is also shown. (B) The mmu_circ_0001359 was fused to the luciferase coding region (pMIR-mmu_circ_0001359) and co-transfected into 293T cells with miR-183-5p mimics, to confirm that mmu_circ_0001359 interacts with miR-183-5p. Both pMIR-mmu_circ_0001359 and miR-183-5p mimic constructs were co-transfected into 293T cells with a control vector, and relative luciferase activity was determined 48 h after co-transfection. Data are mean ± SD. **p < 0.01. (C) Complementary sequences in miR-183-5p and the 3′ UTR of FoxO1 mRNA were obtained using publicly available algorithms. The mutated version of the FoxO1 3′ UTR is also shown. (D) The 3′ UTR of FoxO1 was fused to the luciferase coding region (pMIR-FoxO1 3′ UTR) and co-transfected into 293T cells with miR-183-5p mimics, to confirm that FoxO1 was the target of miR-183-5p. The pMIR-FoxO1 3′ UTR and miR-183-5p mimic constructs were co-transfected into 293T cells with a control vector, and relative luciferase activity was determined 48 h after co-transfection. Data are mean ± SD. **p < 0.01.