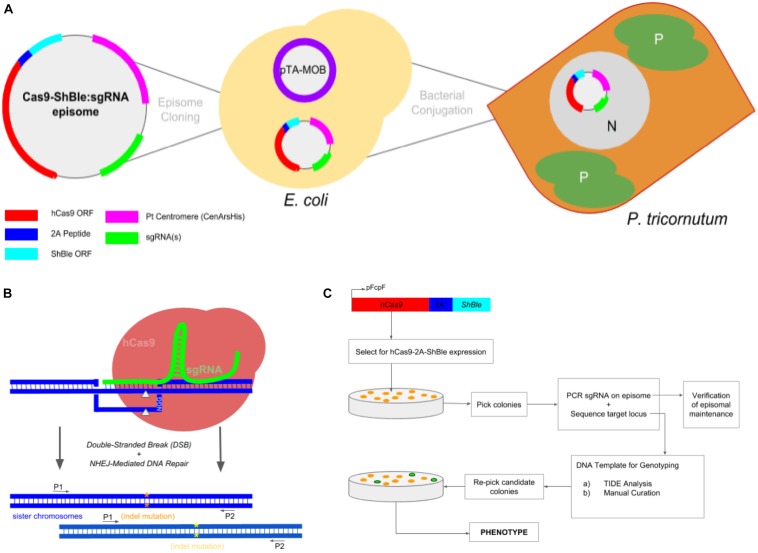
FIGURE 1.
Experimental workflow to produce Phaeodactylum mutants via conjugation-based delivery of a CRISPR-Cas9 episome. (A) A cartoon schematic of Cas9-Sh ble:sgRNA episome cloning to E. coli cells harboring the pTA-MOB plasmid and followed by bacterial-conjugation transformation of the episome to Phaeodactylum. Phaeodactylum cells maintain and replicate the Cas9-Sh ble:sgRNA episome as their native chromosomes in the nucleus (N). The diatom plastids (P) and cytoplasm (C) are labeled. Here, Cas9 is transcriptionally fused to sh ble and can therefore be selected for after transformation with the antibiotic phleomycin. (B) Episomal expression of Cas9 and sgRNA(s) and target mutagenesis of a diploid organism. The sgRNA(s) guides Cas9 to genomic target(s), and then Cas9 induces a double-stranded break (gray arrows). Phaeodactylum, being a diploid eukaryote, will contain two distinct NHEJ-mediated mutations (orange and yellow asterisks) between the sister chromosomes. (C) Genotyping and cell line picking workflow after colony selection for Cas9-Sh ble. A “double picking” strategy was used by which cell lines were picked and target locus sequenced using two primers. After genotyping, the colony can be re-picked for further cell line analysis.