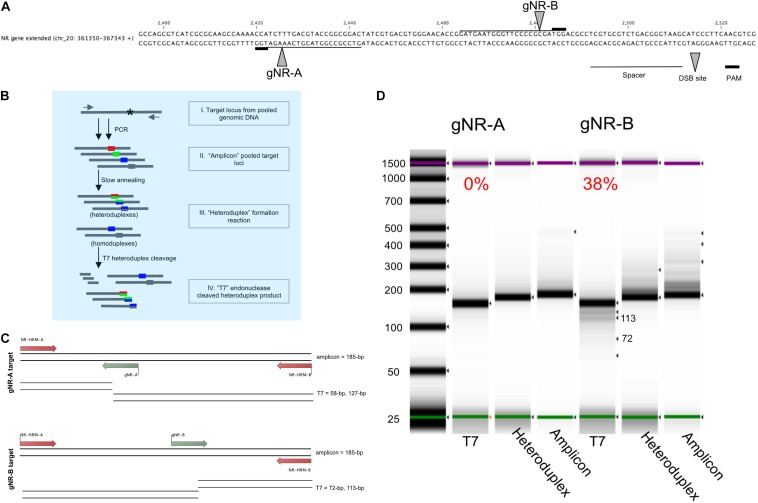
FIGURE 2.
Pooled-diatom transformant T7 assay screen for in vivo Cas9 activity to compare the mutagenesis efficiency between two sgRNAs designed to target the nitrate reductase (Phatr3_J54983) gene. (A) Two sgRNAs were designed within the Phaeodactylum genomic region displayed. The spacer sequence (line), PAM site (bold line) and Cas9 cut site (grew arrow) are defined for sgRNA gNR-A and gNR-B. (B) The T7 heteroduplex assay was used to quantify the efficiency of in vivo Cas9 activity for each sgRNA for a population of pooled-Phaeodactylum transformants. This chart demonstrates how the T7 heteroduplex assay can produce and cleave mismatched amplicon products(I). The pool of target loci amplicons (II) is denatured and slowly annealed to form mismatched double-stranded DNA, or heteroduplexes (III). The heteroduplexes are then digested by T7 endonuclease I (New England Biolabs). Heteroduplexes are subsequently cleaved by T7 while homoduplexes remain full length. (C) The expected T7 cleavage pattern for gNR-A and gNR-B associated amplicons. The size of the “amplicon” input is 185-bp. For gNR-A, the expected “T7” products are ∼185-bp (homoduplex), 127-bp and 58-bp. For gNR-B, ∼185-bp (homoduplex), 113-bp and 72-bp. (D) The three products, “amplicon,” “heteroduplex,” and “T7” were size analyzed on a 4200 TapeStation System (Agilent, Santa Clara, CA, United States). The T7 cleavage efficiency is displayed above each “T7” lane in the gel that represents the mutagenesis efficiency of each sgRNA for all Phaeodactylum cell lines. Each band detected by the software is indicated by a black arrow. The expected “T7” 113-bp and 72-bp bands are labeled.