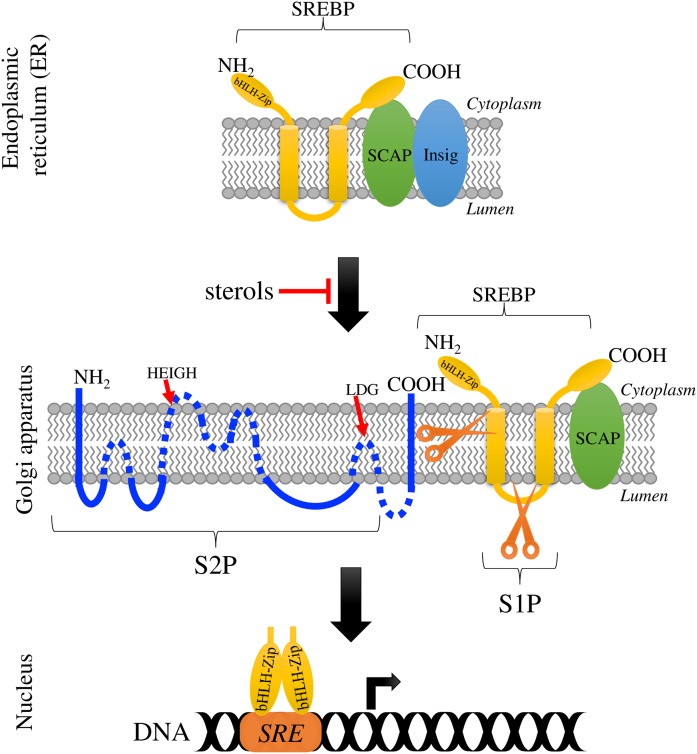
Fig. 1.
The SREBP pathway. Scheme of the mammalian SREBP pathway [adapted from (67)]. With sufficient sterol, the SREBP-SCAP complex is retained at the ER membrane due to SCAP interacting with Insig; when sterol levels decrease, the SREBP-SCAP complex is transported to the Golgi apparatus. In the Golgi apparatus, SREBP undergoes two sequential proteolytic cleavages by S1P and S2P. First, S1P (subtilisin-related serine protease) cleaves SREBP at the hydrophilic loop projected into the lumen of the Golgi apparatus, and then S2P (metallopeptidase) cuts SREBP within the first TM segment. This releases the N-terminal domain of SREBP (the activated transcription factor), which travels to the nucleus to regulate gene transcription of target genes, including genes required for the synthesis and uptake of sterols and other lipids. bHLH-ZIP represents the N-terminal domain of SREBP, containing the bHLH leucine zipper motif, and SRE denotes the sterol regulatory element. The illustrated S2P membrane topology was based on the results of Zelenski et al. (42), and conserved residues in the motifs HEIGH and LDG involved in the coordination of the zinc atom are shown.