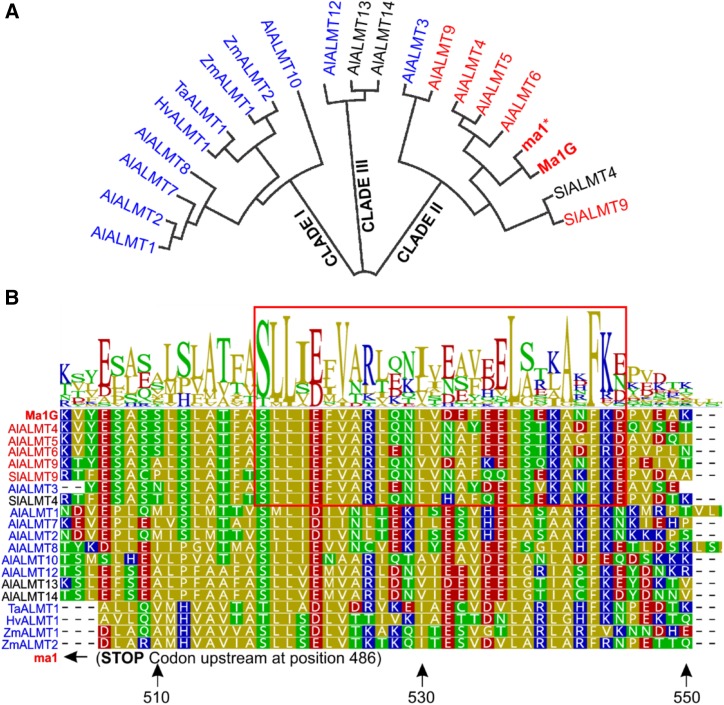
Figure 6.
The conserved C-terminal domain of Ma1 and other tonoplast-localized ALMT members. A, Phylogenetic analysis of Ma1G, ma1, and the 13 ALMT Arabidopsis members, two tomato, two maize (Zea mays), one barley (Hordeum vulgare), and one wheat ALMT members based on amino acid sequence alignment by the neighbor-joining method using Geneious Tree Builder (https://www.geneious.com/). AtALMTs cluster into three groups: I, II, and III. Clade I contains ALMTs known to localize to the PM (colored in blue). Clade II contains ALMTs know to localize to the tonoplast (colored in red), including Ma1G and ma1. B, Sequence alignment and logo illustrating the degree of conservation in a C-terminal domain of the ALMTs described in A. The residue positions marked by the arrows at the bottom of the alignment are relative to the Ma1G sequence. The early stop codon in ma1 is located 17 residues upstream of the aligned sequences. The red box highlights the amino acid region 519 to 545 for the clade II ALMTs.