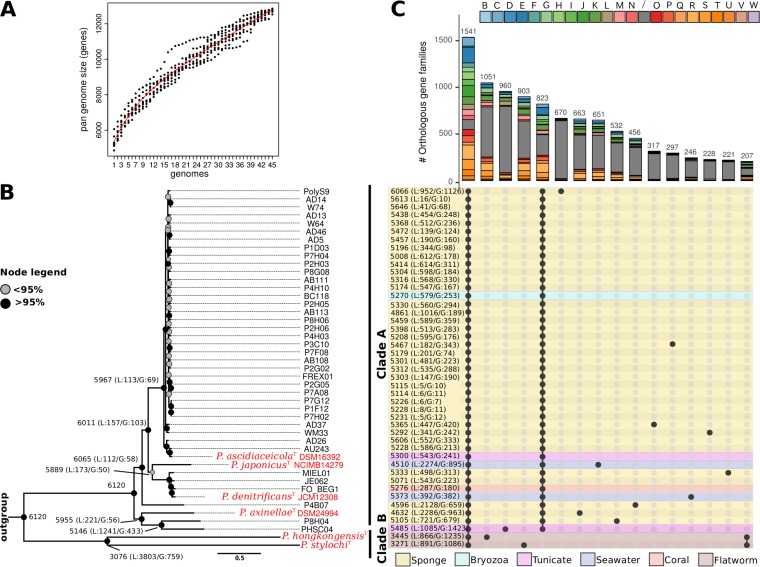
FIG 1.
Genomic composition of the Pseudovibrio genus. (A) A resampling procedure was used to estimate the dimension of the pan-genome of the genus. (B) After inclusion of the outgroup Labrenzia alexandrii DFL-11 (GCA_000158095.2) in the homologous gene family identification, a phylogenomic tree was constructed selecting only those genes within the core genomes that could give a well-resolved tree topology and that did not show signs of recombination. Bootstrapping with 100 replicates was performed to obtain approximate Bayes branch support values. (C) Distribution of the homologous gene families among the Pseudovibrio strains, visualized using the UpSetR package (100). The histogram indicates the number of orthologous gene families, with their distribution profiles indicated by filled circles in the matrix at the bottom. Bars of the histogram are colored according to the COG categories to which the orthologous families have been assigned. The panel reporting the distribution profile of the orthologous families is colored according to the source of isolation of the strains. Only orthologous gene families containing > 200 genes are reported.