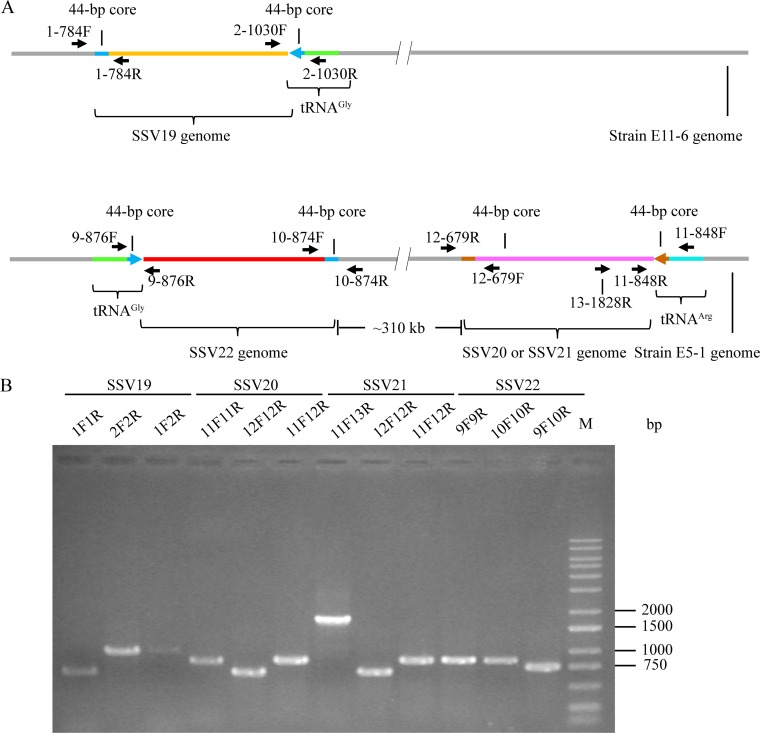
FIG 7.
Detection of the integration of SSV19-22 DNA into their host genomes. (A) A diagram showing the sites of integration for SSV19-22 as well as the positions of primers used in the analysis of viral integration. The tRNA genes, in which integration occurred, and primer pairs flanking the attP sites are indicated. (B) Occupancy of the integration sites on the host chromosomes. DNA extracted from virus-infected host cells was amplified using specific primer pairs for integrated and unoccupied sites. PCR products were loaded onto a 1.0% agarose gel. After electrophoresis, the gel was photographed under UV light. The expected sizes of the PCR products were 784 bp (1F/1R), 1,030 bp (2F/2R), 1,033 bp (1F/2R), 876 bp (9F/9R), 874 bp (10F/10R), 758 bp (9F/10R), 848 bp (11F/11R), 679 bp (12F/12R), 860 bp (11F12R), and 1,828 bp (11F/13R), respectively. M, molecular weight standards.