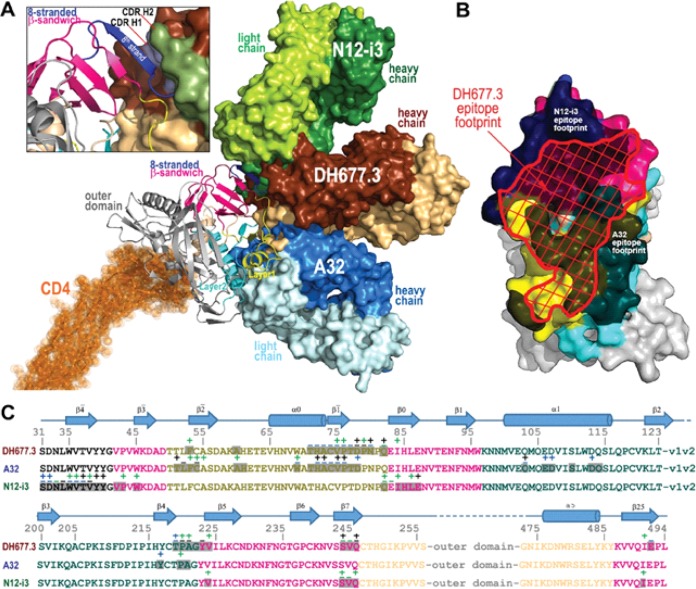
FIG 6.
Recognition of HIV-1 Env by DH677.3 and other cluster A MAbs. (A) The overlay of DH677.3 and cluster A MAbs A32 and N12-i3 (C11-like) bound to the gp120 core. Crystal structures of the gp120 antigen in complex with the Fab of DH677.3, A32 (PDB code 4YC2), and N12-i3 (PDB code 5W4L), superimposed based on gp120. The d1 and d2 domains of the target cell receptor CD4 were added to replace peptide mimetic M48U1 of the DH677.3 Fab-gp12093TH057 coree-M48U1 complex. Molecular surfaces are displayed over Fab molecules and colored in lighter and darker shades of brown, blue, and green for the heavy and light chains of DH677.3, A32, and N12-i3, respectively. A blow-up view shows details of the DH677.3 interaction with the 8-stranded β-sandwich of the gp120 inner domain. The 8th strand (colored in blue) formed by the 11 N-terminal residues of gp120 in the N12-i3 bound conformation (PDB entry 5W4L) was modeled into the DH677.3 Fab-gp12093TH057 coree-M48U1 complex. CDR H1 and H2 of DH677.3 are colored light blue and dark green, respectively. (B and C) Comparison of DH677.3, A32, and N12-i3 epitope footprints. (B) The DH677.3 epitope footprint (shown in red) is plotted on the gp120 surface with layers colored, as described for Fig. 1, with the A32 and N12-i2 epitope footprints shown in black. (C) DH677.3, A32, and N12-i3 gp120 contact residues are mapped onto the gp120 sequence. Side-chain (+) and main-chain (−) contact residues are colored green for hydrophobic, blue for hydrophilic, and black for both, as determined by a 5-Å cutoff value over the corresponding sequence. Buried surface residues, as determined by PISA, are shaded. The DH677.3 epitope footprint overlays with the epitopes of both A32 and N12-i3.