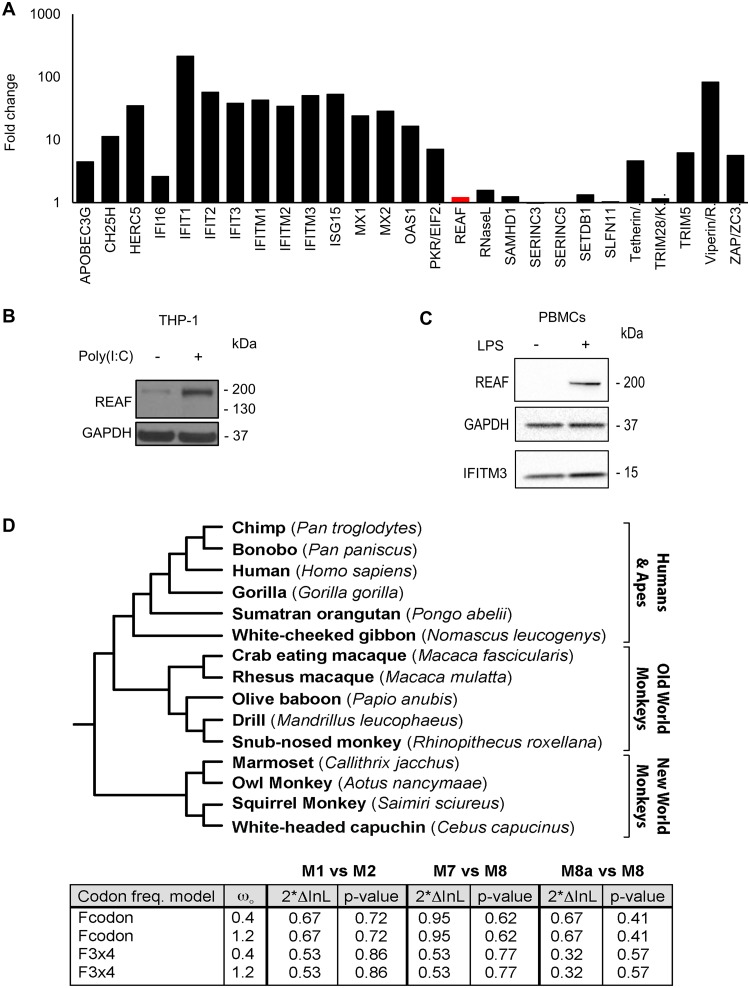
FIG 4.
REAF is not IFN stimulated or under positive selection. (A) RNA microarray analysis determined changes in REAF mRNA compared to other antiviral factors in MDMs treated with IFN-α (500 IU/ml). (B) REAF protein in PMA-differentiated THP-1 cells after poly(I·C) treatment for 48 h. GAPDH was a loading control. (C) REAF protein in PBMCs after LPS treatment. GAPDH was a loading control, and IFITM3 was a positive control for LPS-induced upregulation. (D) REAF DNA sequences from 15 extant primate species (tree length, 0.2 substitutions per site along all branches of the phylogeny) (top) were analyzed using the PAML package for signatures of positive natural selection (bottom). Initial seed values for ω (ωO) and different codon frequency models were used in the maximum-likelihood simulation. Twice the differences in the natural logarithms of the likelihoods (2*InL) of the two models were calculated and evaluated using the chi-squared critical value. The P value indicates the confidence with which the null model (M1, M7, or M8a) can be rejected in favor of the model of positive selection (M2 or M8).