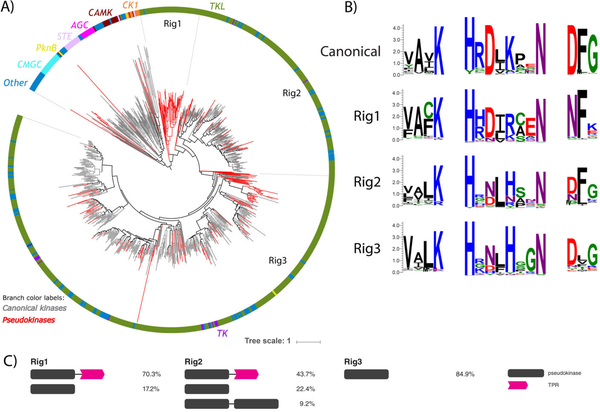
Fig. 5.
Rhizophagus irregularis-specific TKL pseudokinase families. (A) Phylogenetic tree of the R. irregularis kinome. Canonical kinase branches are colored in gray and pseudokinases in red. Major kinase groups are labelled using different colors in the outer circle. The 3 major R. irregularis specific pseudokinase families are labelled as Rig1, Rig2 and Rig3. (B) Sequence logos of catalytic motifs for Rig1, Rig2, and Rig3 pseudokinase families. (C) The most common domain structures observed in Rig pseudokinase families are shown (occurring >5%), with frequencies of each domain structure indicated.