Figure 2.
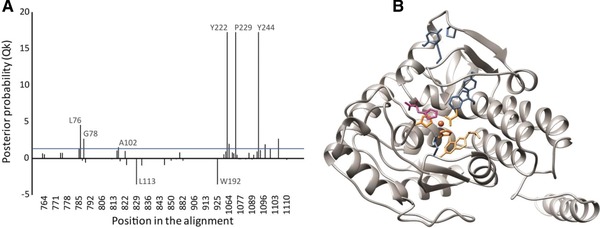
Prediction of substrate‐determining residues based on functional divergence type‐II analysis of tryptophan and phenylalanine hydroxylase, and their position in Cupriavidus taiwanensis modeled structure: (A) posterior probability (Qk) of the homologous sites, a threshold line of Qk = 0.9 is indicated in the graph; (B) predicted amino acids (blue) are mapped in the structure, phenylalanine (magenta) is present at the active site, as well as the cofactor, 2‐His‐1‐caboxylate facial triad and the iron molecule (orange).
