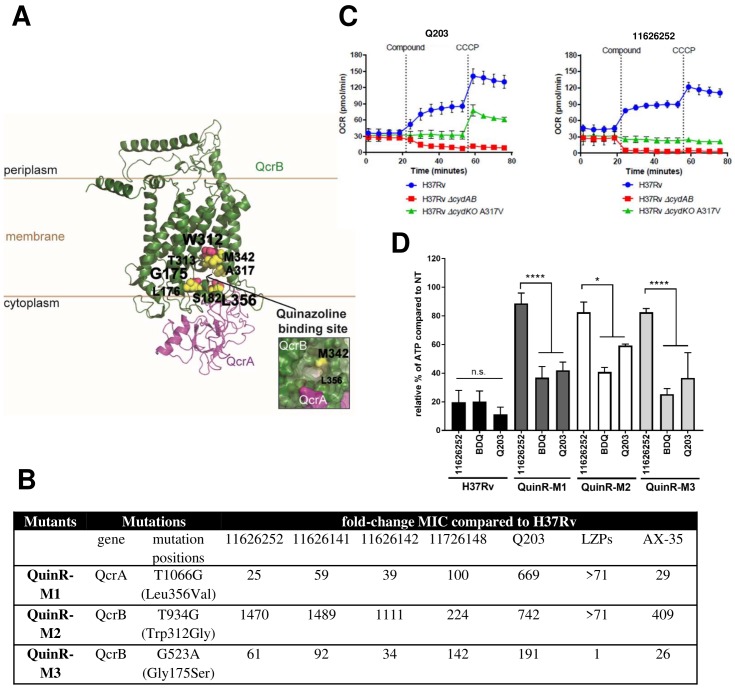
Fig 3. Quinazoline derivatives target cytochrome bc1.
(A) QcrA and QcrB of M. tb were modelled using the homology modelling webserver (Swissmodel) and homology based on chains A and M from the CryoEM structure of the III2IV2 respiratory supercomplex (PDB code 6HWH) from M. smegmatis. Clustering of mutations associated with resistance to quinazoline derivatives, AX-35, Q203, and LPZs occurs around the quinol oxidation site of QcrB and QcrA. (B) Cross-resistance of QuinR mutants to QcrB inhibitors. The MIC of the quinazoline derivatives, Q203, LPZs and AX-35 was determined and expressed as the fold-change of MIC of each compound compared to M. tb H37Rv. (C) Bioenergetic analysis of M. tb upon Q203 and 11626252 treatment followed by the uncoupler CCCP to stimulate maximum respiration. The oxygen consumption rates of wild-type H37Rv, H37Rv ΔcydAB, and H37Rv Δcyd KO with a QcrB (A317V) mutation (Q203 resistance SNP) was monitored as described in the Material and methods section. (D) Intracellular ATP levels in H37Rv and quinazoline resistant strains were measured in the absence and presence of BDQ, Q203, and 11626252 at 2.5× MIC after 24 h using BacTiterGlo (Promega). Data from two independent experiments are presented as mean ± SD. Statistical analysis was performed using two-way analysis of variance (ANOVA) with Tukey’s multiple-comparison tests (*, P < 0.03; ****, P < 0.0001).