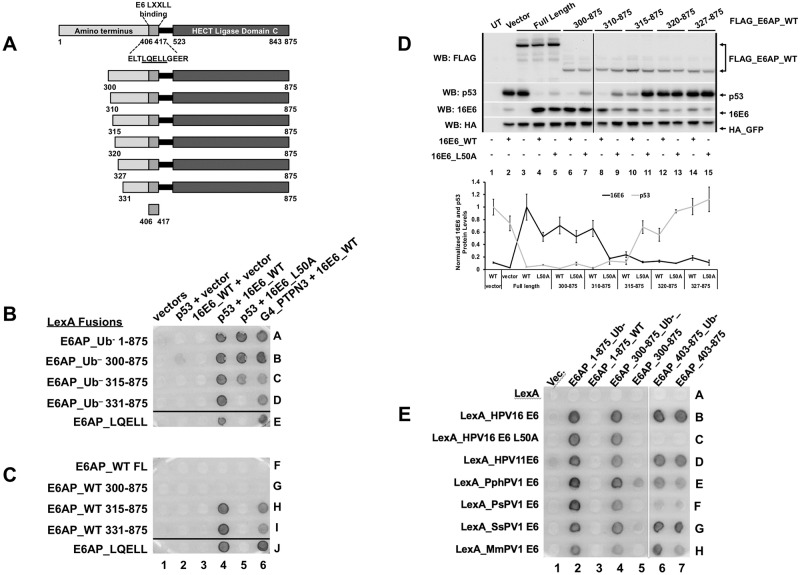
Fig 3. The amino terminal region of E6AP is required for 16E6 degradation of p53.
(A) Schematic of E6AP amino terminal truncations. Previously described E6 LQELL binding region is between amino acids 403 and 416, as depicted. Each E6AP amino terminal truncation retains the E6 LQELL binding region and the E6AP carboxy terminal HECT ligase domain. (B) Ability of 16E6_L50A to bind to E6AP requires E6AP residues 315–331. Yeast strains expressing the LexA DNA binding domain fused to ubiquitin ligase dead (Ub–) E6AP full length (FL), amino terminal E6AP truncations, or the isolated E6AP LQELL peptide (406–417). These bait yeasts were mated to yeast strains co-expressing p53, 16E6_WT, 16E6_L50A, or transactivator Gal4 (G4) fused to the PDZ protein PTPN3 as indicated. Positive controls include 16E6_WT co-expressed with p53 to ensure E6AP expression and G4-PTPN3 co-expressed with 16E6_WT to ensure 16E6_WT expression. Ability of 16E6_L50A to bind E6AP_Ub- and recruit p53 is lost when E6AP is truncated from residue 315 to residue 331. Horizontal black line indicates removal of an irrelevant sample. (C) Ability of E6 to stimulate ubiquitin ligase activity in the complex with p53 and PTPN3 requires E6AP amino acids 310–315. Bait yeast were transfected with the LexA DNA binding domain fused to WT E6AP containing the same truncation endpoints as described in B. These yeast strains were mated to the same yeast as described in B and diploids selected. Upon truncation of E6AP from amino acid 300 to 315, the interaction of p53 and PTPN3 with E6 was restored as indicated by the appearance of blue dye. Horizontal black line indicates removal of irrelevant samples. (D) Requirement of E6AP amino acids 310–320 for E6 ability to initiate p53 degradation in the presence of E6AP_WT is recapitulated in E6AP-null 8B9 cells. Plasmids encoding the indicated FLAG_E6AP_WT truncations (1.25 ug), human p53 (0.5 ug), HA-GFP (0.01 ug), and either untagged 16E6_WT or 16E6_L50A (2 ug), as indicated, were transiently transfected into murine E6AP-null 8B9 cells and p53 and E6 expression were analyzed by western blot. 16E6_WT requires E6AP amino acids 315–320 to initiate E6AP-mediated degradation of p53 while 16E6_L50A requires E6AP amino acids 310–315. A single representative blot is shown. Vertical black line indicates removal of an irrelevant sample. UT = untransfected. Quantitation of protein expression from triplicate experiments as shown below the blots. E6AP stabilization of 16E6 (black line) is not required for p53 (gray line) degradation by the E6-E6AP complex. p53 levels and E6 levels are normalized to co-transfected HA_GFP. HA_GFP normalized E6 expression levels are further normalized to 16E6_WT protein levels in the presence of full length E6AP (lane 4 in panel D). HA_GFP normalized p53 protein expression levels are normalized to p53 levels in the presence of 16E6_WT with no co-expressed E6AP protein (lane 2 in panel D). The means of triplicate independent experiments ± standard errors are shown. E. Diverse E6 proteins require E6AP region 300–403 for ubiquitin ligase function. The indicated LexA_E6 fusions from Fig 1 are expressed together with transactivator fusions of E6AP active for Ubiquitin ligase activity (WT) or Ubiquitin minus (Ub-) in columns, either full length E6AP_1–875, or amino-terminal truncations 300–875 or 403–875. White vertical line indicates the division from 2 matched plates.