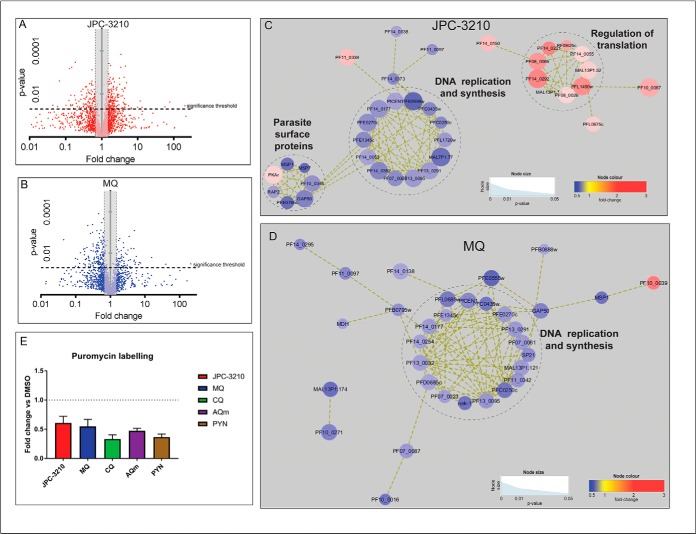
Fig. 8.
Proteomics analysis of dysregulated protein abundances following JPC-3210, and MQ treatment. A–B, Volcano plot of differential protein abundance following treatment. Proteins identified in at least three independent experiments following treatment with 1 μm (A) JPC-3210 (2014 proteins, indicated by red color dots) or (B) MQ (2014 proteins, blue dots) for 5 h. Proteins above the significance threshold (p value < 0.05) and outside the gray shaded area (fold change ≥ 1.5) were considered significant. C–D, Network analysis of trophozoite-stage parasite proteins perturbed following treatment with (C) JPC-3210, and (D) MQ. The network analysis was built using the STRINGdb interaction network analysis output (connectivity was based on experimental, database and co-expression evidence with a minimum interaction score of 0.7) in Cytoscape 3.6 with the ClusterONE algorithm. Node size represents p value and node color represents fold-change from at least three independent replicates. E, Puromycin labeling of trophozoite stage 3D7 parasites treated with 1 μm JPC-3210 (red), CQ (green), AQm (purple), PYN (gold) or a DMSO control for 5 h. Bars represent the ratio of labeled puromycin for the respective compounds relative to the DMSO control ± the S.E. from three independent replicates.