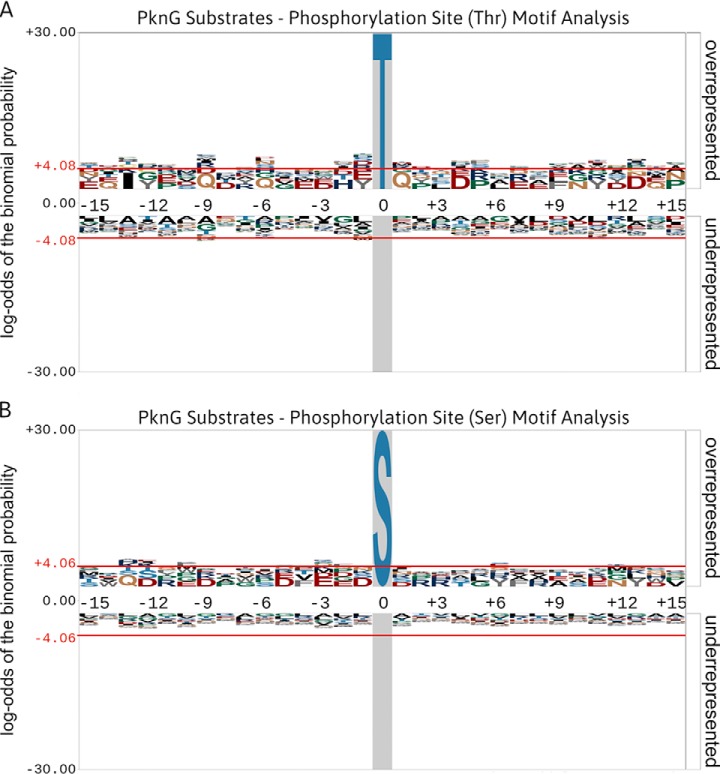
Fig. 6.
Phosphorylation site motif analyses of PknG substrates generated using pLogo (68). (A) An in silico motif analysis of Thr-phosphorylated peptides (n = 26) derived from PknG substrates. (B) An in silico motif analysis of Ser-phosphorylated peptides (n = 12) derived from PknG substrates. Phosphorylation site motifs were analyzed using a foreground composed of available sequence windows extracted from the studies cited in Table II. The M. tuberculosis (strain ATCC 25618/H37Rv) proteome was used as the background database. The red horizontal lines (± 4.06) illustrate the relative statistical significance (p value ≤ 0.05, after Bonferroni correction) of residues flanking the central phosphorylation site. Overrepresented residues are above the midline, whereas underrepresented residues are below the midline. No distinct motifs were observed for phosphorylation on either Thr or Ser sites.