Figure 9.
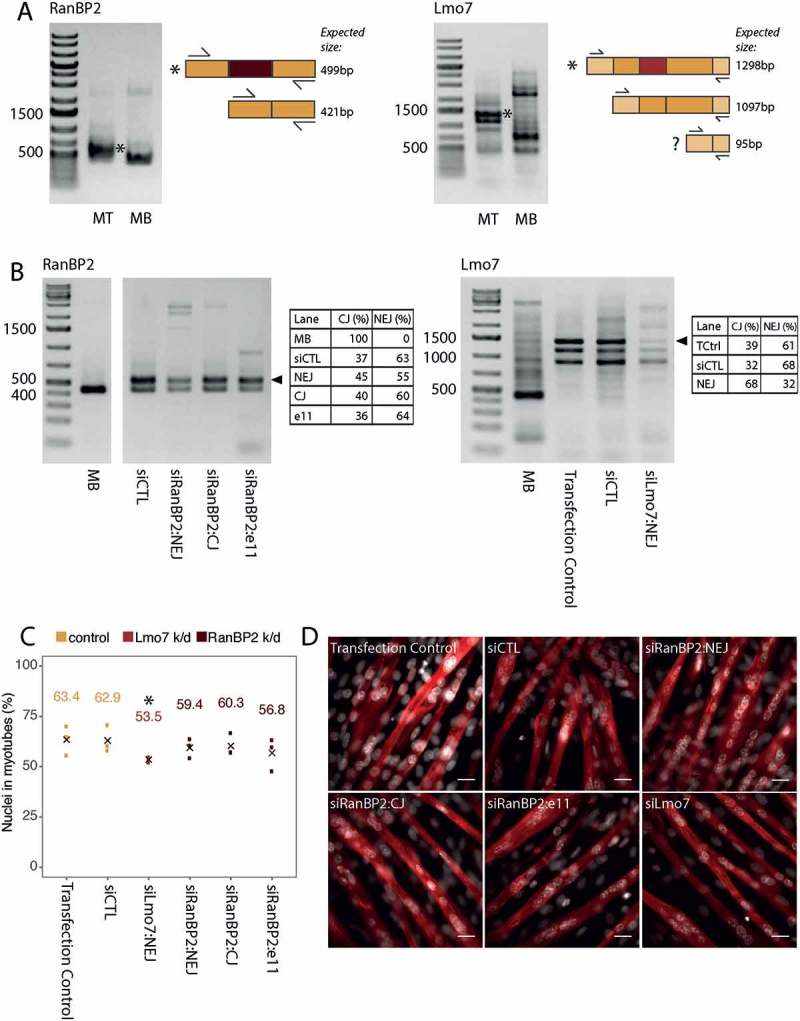
Functional validation of RanBP2 and Lmo7 novel splice variants in mouse C2C12 myogenesis. (a) RT-PCR for the novel myotube-specific exons reveals that they are spliced into mRNA produced in C2C12 mouse cells differentiated into myotubes (* marks novel splice form). Adjacent diagrams indicate the primer design and expected amplicon sizes. Note that for Lmo7 a third splice isoform is expected (marked with a question mark), however we don’t observe this in our gels (MB: myoblast, MT: myotube). (b) siRNAs against either the novel exon junction (NEJ), or canonical exon junction (CJ), or against a separate exon (e11), result in reduced mRNA expression of the specific RanBP2/Lmo7 splice variants. Novel splice variants are marked by an arrowhead. siCTL refers to treatment with a non-target control siRNA. Transfection control refers to treatment with the transfection reagents alone, but no siRNA. Tables next to each gel show the proportion of the novel and canonical isoforms in each quantifiable lane, as measured using the in-built gel analyzer tool in Fiji. (c) Quantification of myogenic index (percentage of nuclei in myotubes) as determined by myosin heavy chain 1 staining (red) in 4 day differentiated myotubes. The numbers above the points represent mean values from three biological replicates, the mean is also indicated with black crosses. * represents a statistically significant difference (p < 0.0001) between knockdown and non-target siRNA control conditions as measured using a Fisher’s exact test on the aggregated replicates. (d) Example fields of view used for determination of myogenic index in (c). Scale bars = 15 μm.
