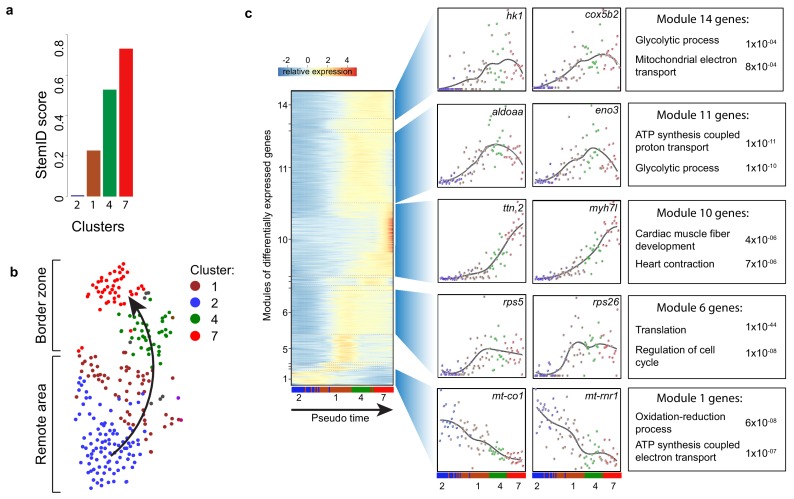
Figure 3. Pseudo time analysis reveals dedifferentiation and metabolic changes in border zone cardiomyocytes.
(a) Bar plot of StemID scores for the cardiomyocyte clusters (clusters #2, 1, 4 and 7) calculated by the formula: number of significant links for each cluster multiplied by the median transcriptome entropy across all cells in a cluster. (b) Cardiomyocyte clusters from adult injured heart. Arrow indicates the dedifferentiation path derived from the StemID scores. (c) Pseudo time analysis. Left; one-dimensional SOM of z-score transformed expression profiles along the differentiation trajectory incurred by StemID analysis. Y-axis represents the fourteen modules with differentially expressed genes. X-axis represents the pseudo time in which the cells were ordered. Middle; expression profiles of representative genes of the major modules. Y-axis shows transcript counts. X-axis represents the pseudo time. Right; Major gene ontology terms derived from all genes expressed in the module with p-values.