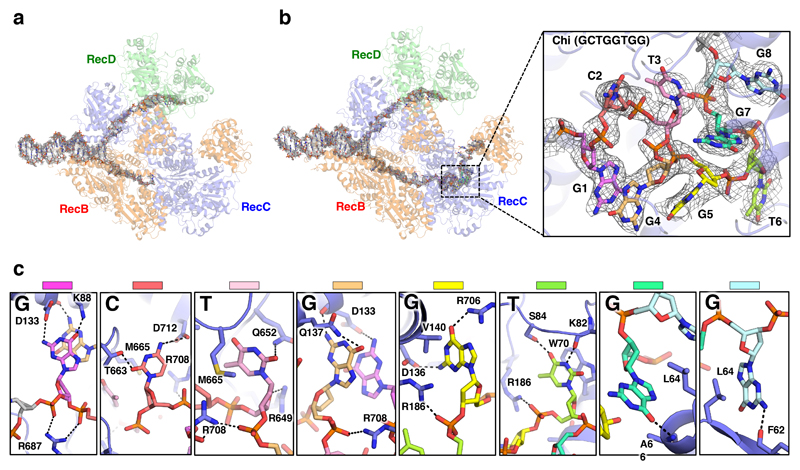
Fig. 2. Details of the Chi-binding site interactions.
a, Density corresponding to the bound DNA fork substrate in the Chi-unrecognised state with the protein complex shown as a faint cartoon. Strong density is observed for the duplex and 5’-tail regions. Density is also observed for the first six bases of the 3’-tail, but is disordered beyond that. b, Density corresponding to the bound DNA fork substrate in the Chi-recognised state. The density is similar to that observed in the Chi-unrecognised state except that additional density is observed for the entire 20 bases of the 3’-tail, including the Chi sequence. Inset, Expanded view of the density (contoured at 2.5 σ) for the eight Chi bases with the Chi bases individually coloured as in panel (c) demonstrating the highly twisted conformation of the DNA. c, Details of the interactions with each base of the Chi sequence. Hydrogen bonds are shown as dotted lines.