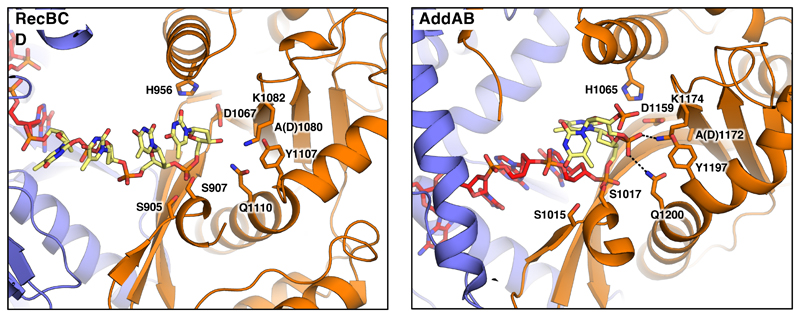
Fig. 5. The nuclease active site in the Chi-bound RecBCD and AddAB complexes.
The nuclease domains of RecBCD and AddAB are shown as cartoons. Key residues participating in DNA binding and catalysis are shown as sticks. Hydrogen bonds are shown as dotted lines. The DNA forks used for both enzymes represent substrates after the final cleavage, with four bases after Chi for RecBCD but just a single base for AddAB (in each case depicted in yellow). The difference in the final cleavage site relates to the distance between the end of the Chi-binding site and the nuclease even though the relative position of the nuclease, relative to RecC or AddB, are similar. The normal cleavage of DNA after Chi in RecBCD would result in a terminal 3’-phosphate but this was not added to the substrate synthesised for the structure determination.