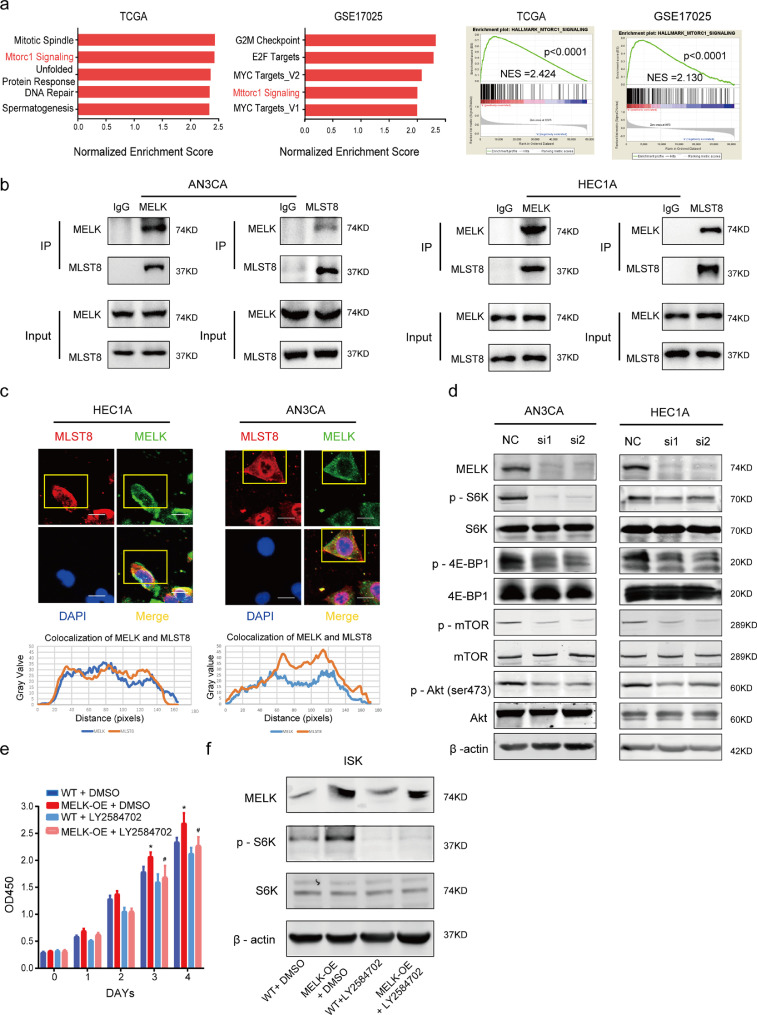
Fig. 5.
MELK activates mTORC1 and −2 by interacting with MLST8. a. GSEA using a hallmark gene set was performed to compare the high MELK expression group and low MELK expression group in both TCGA and GEO databases. MELK expression magnitude in the databases was ordered from low to high: the first 30% of samples were chosen as the low-expression group, and the last 30% of samples were regarded as the high-expression group. Top significant pathways are listed in the left panel, and the mTORC1 signaling pathway was enriched (right panel). b. Co-IP assays were conducted to detect the interactions between MELK and MLST8 in both HEC1A and AN3CA cell lines. c. Immunofluorescence assays were conducted to verify the colocalization of MELK and MLST8. The curves below represent the colocalization of MELK with MLST8, the curves were constructed in the ImageJ software. d. Western blotting analysis of MELK and phosphorylated and total S6K, 4E-BP1, mTOR, and AKT during treatment with MELK-siRNA in both cell lines. β-Actin served as the loading control. e. MELK was artificially overexpressed (MELK-OE) in ISK cells and LY2584702, the S6K inhibitor, was used to treat the MELK-OE and wild type (WT) groups to evaluate the cell proliferation. DMSO is a control. Values are means ± SD, *p < 0.05 (student's t-test) as compared with group “WT+DMSO”; #p < 0.05 (student's t-test) as compared with group MELK-OE+DMSO, n = 3. f. Western blotting analysis of MELK, phosphorylated P70S6K, and total P70S6K according to (e). On the fourth day of treatment with LY2584702, the protein was collected.