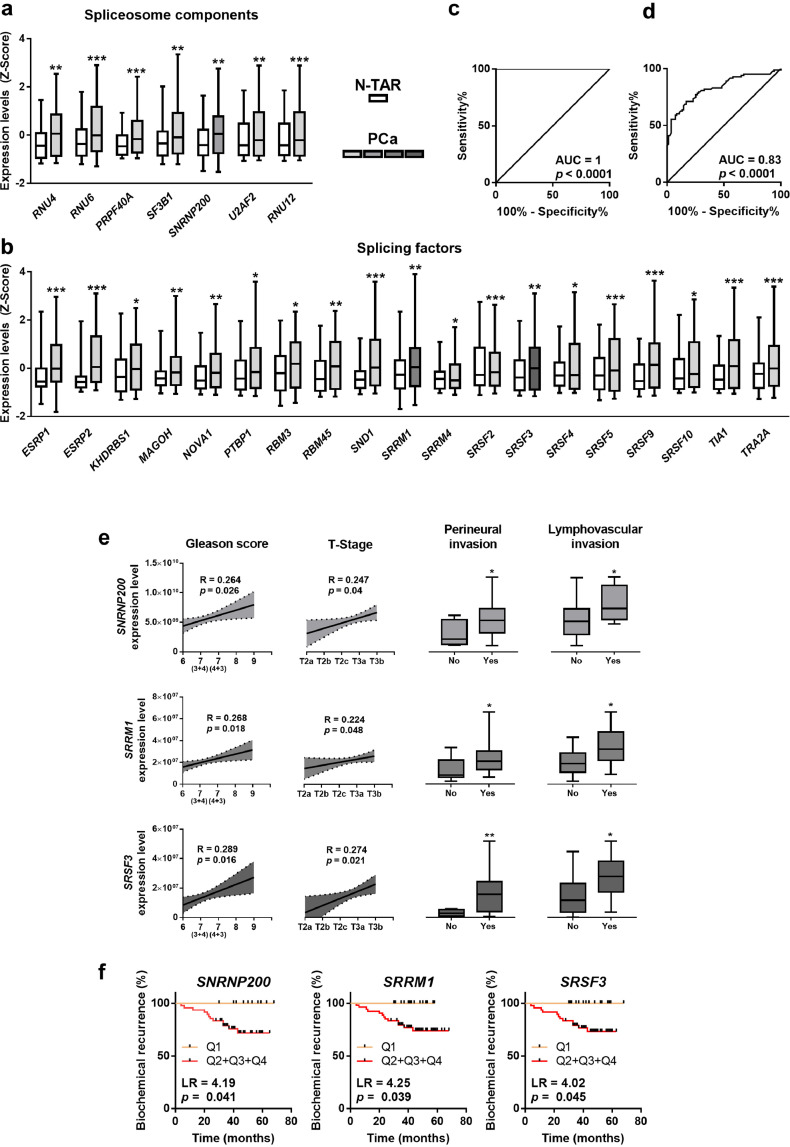
Fig. 1.
Expression of spliceosome components and splicing factors in prostate cancer (PCa) samples. (a–b) Comparison of mRNA levels of spliceosome components (a) and splicing factors (b) between formalin-fixed paraffin embedded (FFPE) samples from PCa samples and non-tumor adjacent regions (N-TAR) (n = 84) determined by a microfluidic-based qPCR array. Data represent the mean ± SEM of mRNA expression levels adjusted by normalization factor (calculated from ACTB and GAPDH expression levels) and standardized by Z-score. c-d) ROC curves of a subset of spliceosome components and splicing factors generated by Random Forest computational algorithm (c) followed by cross validation analysis (d) to distinguish between tumor and N-TAR samples. e) Association between the expression levels of selected spliceosome components and splicing factors (SNRNP200, SRRM1 and SRSF3) and clinical parameters (Gleason score, T-Stage, perineural and lymphovascular invasion) in the same cohort of FFPE samples (n = 84). Correlations are represented by mean (connecting line) and error bands (pointed line) of expression levels. Data of associations represent the mean ± SEM of mRNA expression levels adjusted by normalization factor (calculated from ACTB and GAPDH expression levels). f) Association between SNRNP200, SRRM1 and SRSF3 expression levels and biochemical PCa recurrence in 67 samples from FFPE cohort (samples from patients who underwent adjuvant radiotherapy were not included), calculated by Log Rank analysis (LR). mRNA levels were determined by a microfluidic-based qPCR array and adjusted by normalization factor calculated from ACTB and GAPDH expression levels. Asterisks (* p < 0.05; ** p < 0.01; *** p < 0.001) indicate statistically significant differences between groups.