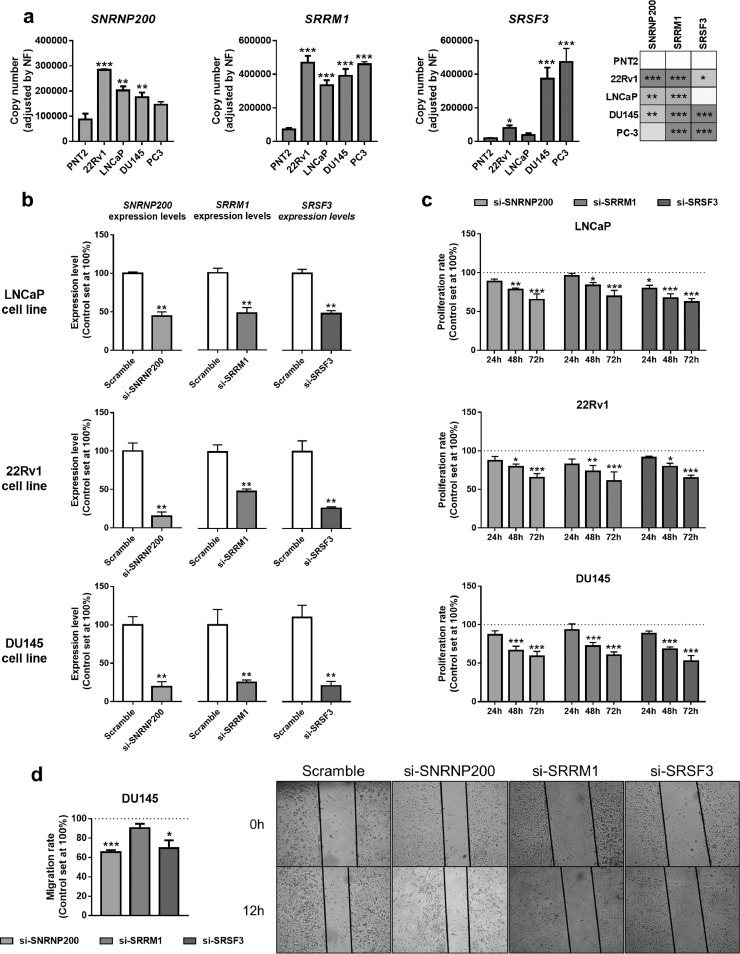
Fig. 4.
Functional consequences of SNRNP200, SRRM1 and SRSF3 silencing in prostate-derived cell lines. a) Comparison of SNRNP200, SRRM1 and SRSF3 expression levels between a non-tumor prostate cell line (PNT2) and PCa cell lines LNCaP, 22Rv1, DU145 and PC-3 (n = 5). mRNA levels were determined by qPCR and adjusted by normalization factor calculated from ACTB and GAPDH expression levels. b) Validation by qPCR of SNRNP200, SRRM1 and SRSF3 silencing (si-SNRNP200, si-SRRM1 and si-SRSF3, respectively). mRNA levels were determined by qPCR and adjusted by normalization factor calculated from ACTB and GAPDH expression levels. Data were represented as percent of scramble cells (mean ± SEM). c) Proliferation rate of LNCaP (upper panel), 22Rv1 (middlepanel) and DU145 (bottom panel) cell lines after 24-, 48- and 72 h of SNRNP200-, SRRM1- and SRSF3-silencing (n = 4). d) Effect of SNRNP200-, SRRM1- and SRSF3-silencing on the migration rate of DU145 cell line was determined by wound-healing assay (12 h; n ≥ 3). Representative images are depicted in right panel. Data were represented as percent of scramble cells (mean ± SEM). Asterisks (* p < 0.05; ** p < 0.01; *** p < 0.001) indicate statistically significant differences between groups.