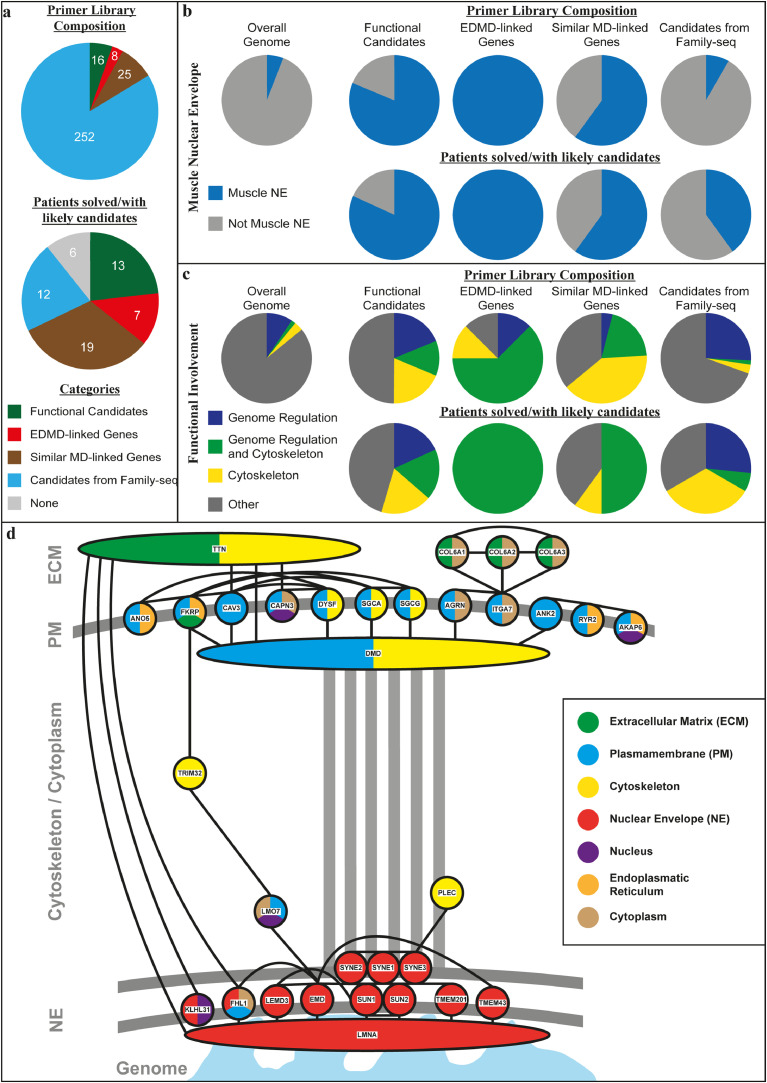
Fig. 2.
Primer library composition and gene ontology (GO) functions/localisations of all candidate genes from the four categories contributing to the primer library construction and for the top candidates identified after primer library sequencing. (a) Composition of the primer library with number of genes from each of the four categories used in its construction (upper panel) and number of patients solved/with likely candidates from the different categories after primer library sequencing (lower panel). (b) Presence in muscle nuclear envelopes for the starting library in comparison to the overall genome (upper panel) and of the remaining candidate genes after primer library sequencing (lower panel) in percent (based on GO-localisation terms and/or experimental evidence from appearance in nuclear envelope proteomics datasets20,28). (c) GO-terms for genome organisation, cytoskeleton, and genome organisation and cytoskeleton combined functions involvement for the starting library in comparison to the overall genome (upper panel) and of the remaining candidate genes after primer library sequencing (lower panel) in percent, showing an enrichment for the combined category in the top candidate alleles. (d) Interactive network of remaining candidate genes after library sequencing based on STRING (Search Tool for the Retrieval of Interacting Genes/Proteins, https://string-db.org/) interactions (high confidence) showing that most candidates are linked to other candidates and that these form connections from the nuclear envelope to the plasma membrane. These connections are consistent with possible mechanotransduction from the extracellular region to the nuclear envelope being the core disrupted function in EDMD. Different described localisations of proteins are displayed by colour-coding (based on GO-terms and/or experimental evidence through identification in muscle nuclear envelope proteomics datasets20).