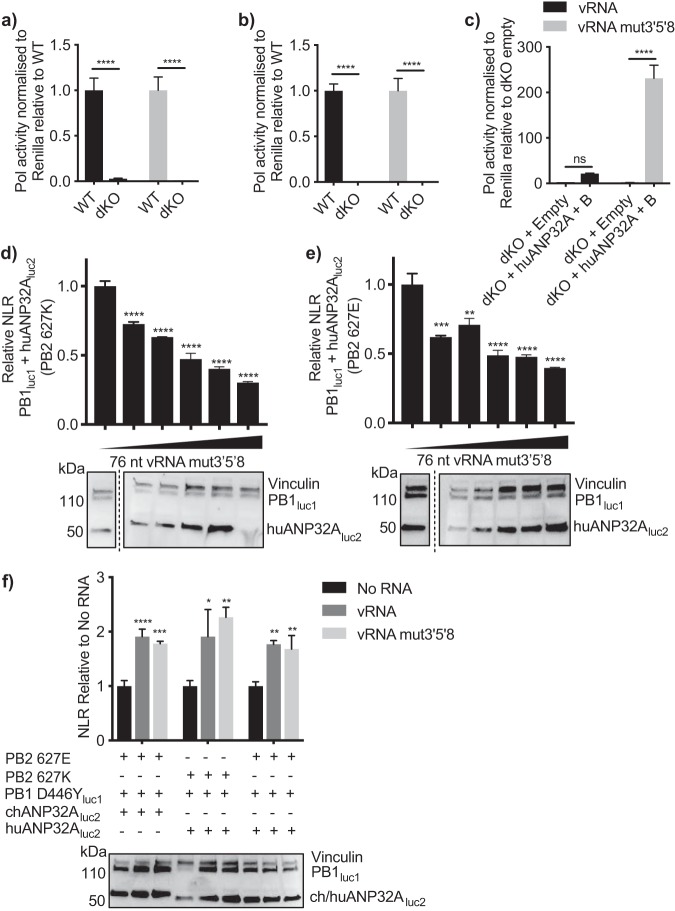
FIG 8.
huANP32A is still required for replication of a vRNA template with 3′5′8 mutations. (a and b) WT or dKO eHAP1 cells were transfected with minigenome assay components containing a polymerase bearing either PB2 627E (a) or PB2 627K (b) and PolI plasmids expressing either wild-type vRNA (black bars) or with the 3′5′8 mutations (gray bars). Results were normalized to Renilla luciferase and presented relative to control cells (WT). (c) eHAP1 dKO cells were transfected with minigenome assay components using only polymerase containing PB2 627E. Either 40 ng empty plasmid or 20 ng huANP32A and 20 ng huANP32B were also expressed. Results were normalized to Renilla luciferase and presented relative to dKO empty plasmid. Statistical significance was assessed by multiple t tests. ns, nonsignificant; **, P < 0.01; ****, P < 0.0001. (d and e) 293T cells were transfected with either PB2 627K (d) or PB2 627E (e) and PB1luc1, PA, and 0, 0.625, 1.25, 2.5, 5, or 10 ng PolI 76-nt vRNA. (f) Cells were transfected with PB1 D446Yluc1, ANP32Aluc2, PA, and either no RNA, 400 ng PolI 76-nt vRNA, or 76-nt vRNA mut3′5′8. Western blotting assay to show expression of tagged plasmids is shown beneath the graphs. Vinculin was used as a loading control. Gaussia luciferase antibody recognizes both luc1 and luc2. Statistical significance was assessed by one-way analysis of variance, and comparisons were made to sample with no added RNA (black bars). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Results representative of three independent experiments.