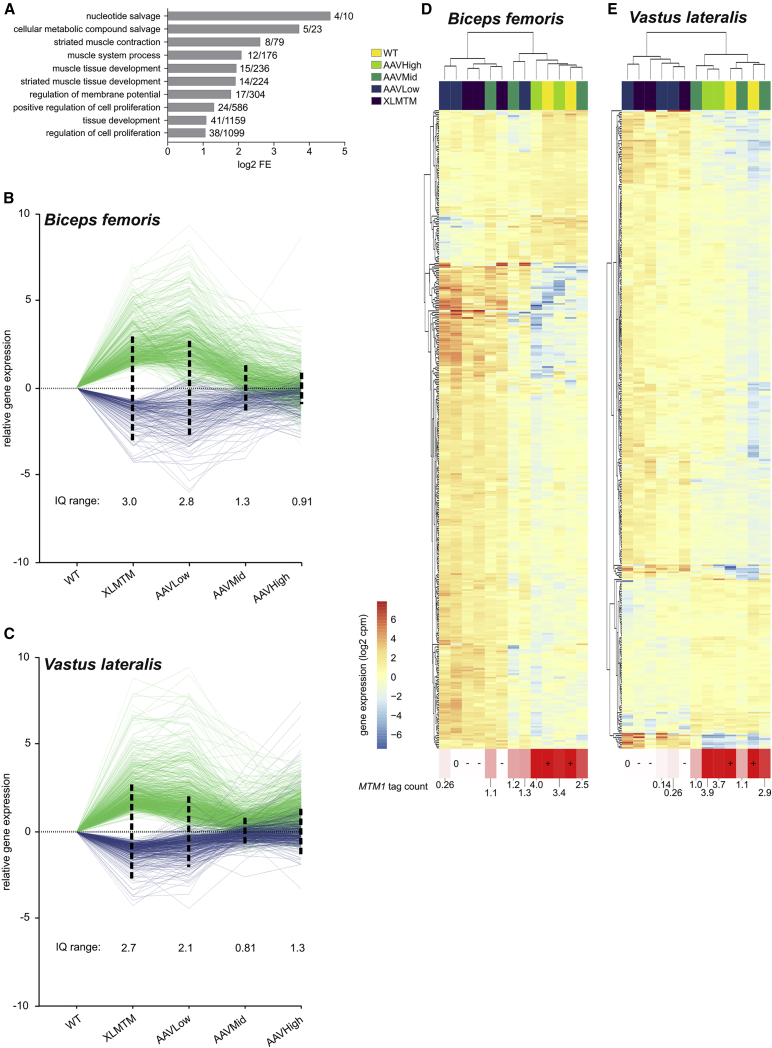
Figure 2.
Remodeling of the XLMTM Transcriptome by rAAV Gene Therapy
(A) Gene ontology analysis on the differentially expressed genes common to XLMTM Biceps and Vastus muscles. For each represented term, the log2 fold enrichment (FE) values are graphically represented, together with the x/y ratio, defined as the number of genes participating to this enrichment (x) over the total number of genes affiliated with the corresponding ontology (y). (B) Relative expression of the differentially expressed genes in the Biceps, expressed in log fold change (log FC) after normalization with healthy controls. The interquartile ranges were determined from positive log FC values (negative log FC were converted into absolute values) and represented as dashed vertical lines pointing in both directions around the y = 0 line. (C) Similar representation for the differentially expressed genes detected in the Vastus muscle. (D and E) Hierarchical clustering generated from log cpm values of the 400 common differentially expressed genes for the Biceps (D) and the Vastus (E). Each column represents one muscle sample, color-coded for treatment group; each row represents one individual gene, color coded for its expression level (log cpm). RNA-seq-based MTM1 mRNA tag counts are indicated under each column.