Table 2.
Hit Compounds Selected for Validation
| ID | Structure | KM (PBG)a (μM) | Vmaxa (nmol/min/mg) |
|---|---|---|---|
| Control | – | 86 ± 5 | 61 ± 2 |
| C5 | 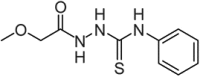 |
81 ± 5 | 55 ± 1** |
| C6 |  |
69 ± 4* | 56 ± 1* |
| C11 |  |
77 ± 5 | 59 ± 1 |
| C17 |  |
81 ± 2 | 58 ± 2 |
Chemical structure of the four hit compounds selected for cellular studies and steady-state enzyme kinetic parameters of HMBS in the presence of the compounds. *p < 0.05 and **p < 0.01 for significant difference compared with the values for the control sample (with 2% DMSO), calculated by unpaired two-tailed t test. Data are presented as mean ± SD.
Effect of the compounds on the enzyme kinetic parameters for HMBS activity, measured at fixed compound concentration (84 μM in 2% DMSO) and variable PBG (0–1 mM) at 37°C, and fitted to Michaelis-Menten kinetics.
