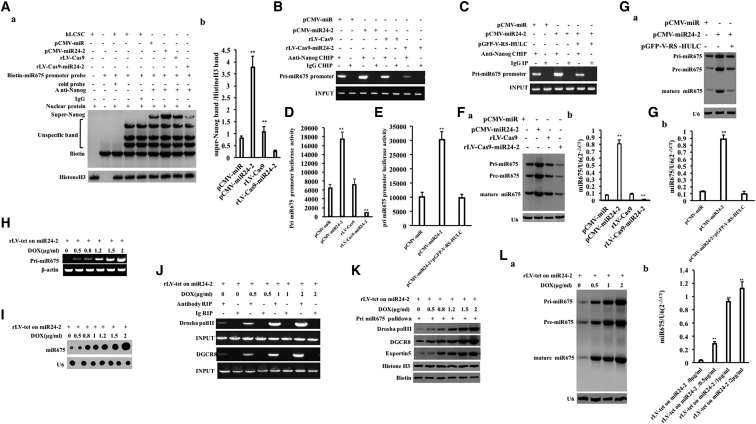
Figure 4.
miR24-2 Promotes Expression and Maturation of miR675
(A) (a) Super-DNA-protein complex gel migration assay using biotin-labeled pri-miR675 promoter cis-element probe and anti-Nanog, anti-Biotin. IgG super-EMSA was used as a negative control. (b) Grayscale scan analysis of positive bands. (B) The ChIP using anti-Nanog. The PCR amplification was carried out using a primer designed according to the pri-miR675 promoter. (C) The ChIP using anti-Nanog. (D) The pEZX-MT-pri-miR675-Luc luciferase reporter activity was assayed. **p < 0.01, *p < 0.05. (E) The pEZX-MT-pri-miR675-Luc luciferase reporter gene activity was detected. (F) (a) Northern blotting was used to detect the miR675. U6 was used as an internal reference gene. (b) Quantitative RT-PCR analysis. (G) (a) Northern blotting was used to detect the miR675. U6 was used as an internal reference gene. (b) Quantitative RT-PCR. (H) The pri-miR675 was analyzed by RT-PCR. β-actin is used as an internal reference. (I) The miR675 was analyzed by dot blotting. U6 is used as an internal reference. (J) RIP with anti-Drosha pol III and anti-DGCR8 was performed. IgG RNA immunoprecipitation was used as a negative control. (K) RNA pulldown was performed. Biotin was used as INPUT and histone H3 was as an internal reference. (L) (a) miR675 was detected by Northern blotting. U6 serves as an internal reference gene. (b) Detection of miR675 by quantitative RT-PCR. U6 was used as an internal reference gene. **p < 0.01, *p < 0.05.