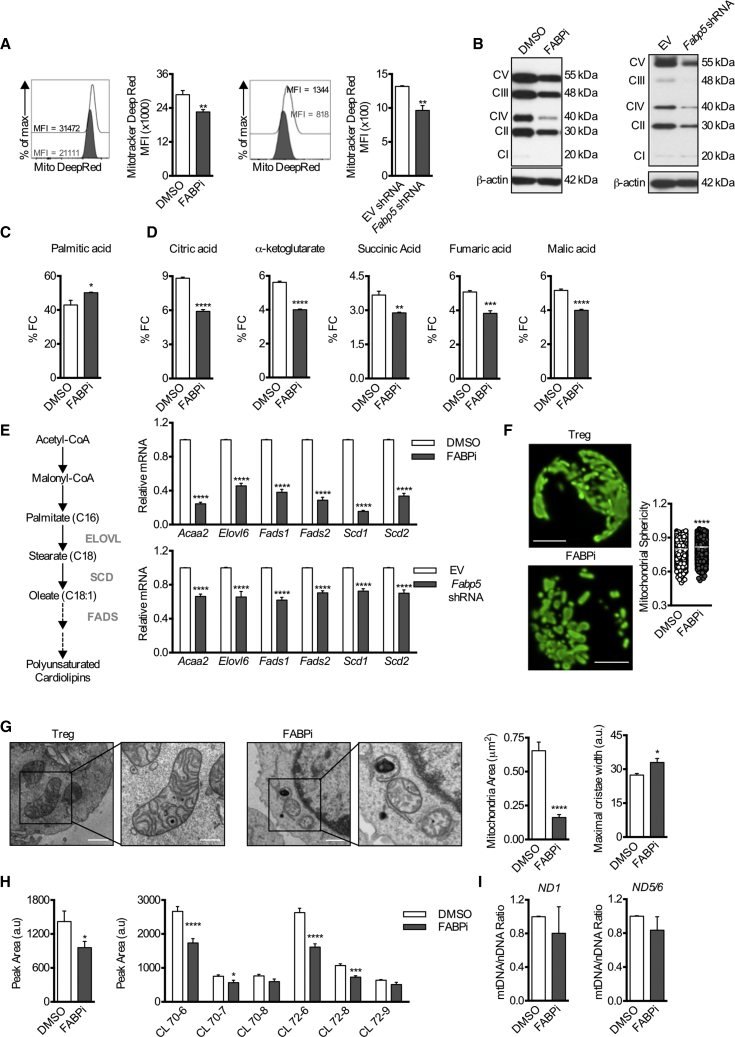
Figure 2.
Impaired OXPHOS, Lipid Metabolism, and Loss of Cristae Structure Underlie Mitochondrial Alterations in Tregs Following FABP5 Inhibition
Naive CD4+ T cells were cultured for 3 days under Treg cell differentiation conditions before overnight BMS309403 treatment or were transduced with lentivirus expressing Fabp5 shRNA.
(A) Representative histogram and mean (±SEM) quantification of MitoTracker deep red dye (n = 4).
(B) Protein expression of the electron transport chain (ETC) complexes. Results represent three independent experiments.
(C) Fractional contribution of 13C-Palmitate-derived carbons to the intracellular palmitate pool in Tregs following acute FABP5 inhibition (n = 4).
(D) Fractional contribution of 13C-Palmitate-derived carbons to intermediates of the TCA cycle in Tregs following acute FABP5 inhibition (n = 4). Results represent two independent experiments.
(E) Schematic of the generation of monounsaturated cardiolipins from acetyl-CoA by the lipid elongation and saturation pathway. Mean (±SEM) expression of genes involved in the lipid metabolism, elongation, and desaturation pathway was assessed by qPCR (n = 4). Results represent two independent experiments.
(F) Tregs were differentiated in vitro from naive CD4+ T cells isolated from PhAM mice for 3 days before overnight BMS309403 treatment. Scale bar, 2 μm. Representative images and quantification of mitochondrial sphericity (n = 4) are shown. Results represent two independent experiments.
(G) Electron micrographs of mitochondria and quantification of mitochondrial area and mitochondrial cristae width from Tregs following acute FABP5 blockade (n = 4). Scale bar, 2 μm in the left panels and 500 nm in the right panels. Results represent two independent experiments.
(H) Left: mean (±SEM) quantification of total cardiolipin content. Right: cardiolipin species in Tregs following overnight treatment with DMSO or BMS309403 (n = 4).
(I) PCR analysis of mitochondrial DNA content (mtDNA/nDNA) in Tregs following overnight treatment with DMSO or BMS309403. Mitochondrial ND1 or ND5/6 were normalized to nuclear gene expression of 18 s. Results represent two independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001. P values were calculated using a two-tailed, unpaired t test (A, C, D, F, left panel, and I) or a two-way ANOVA with Bonferroni correction (E and H, right panel).