Abstract
Background & Aims
Patients with primary biliary cholangitis (PBC) exhibit reduced AE2/SLC4A2 gene expression in the liver and peripheral blood mononuclear cells (PBMCs). AE2 encodes a Cl–/HCO3– exchanger involved in biliary bicarbonate secretion and intracellular pH regulation. Reduced AE2 expression in PBC may be pathogenic, as Ae2-knockout mice reproduce characteristic PBC features. Herein, we aimed to identify CpG-methylation abnormalities in AE2 promoter regions that might contribute to the reduced gene transcription in PBC livers and PBMCs.
Methods
CpG-cytosine methylation rates were interrogated at 1-base pair resolution in upstream and alternate AE2 promoter regions through pyrosequencing of bisulphite-modified genomic DNA from liver specimens and PBMCs. AE2a and alternative AE2b1 and AE2b2 mRNA levels were measured by real-time PCR. Human lymphoblastoid-T2 cells were treated with 5-aza-2´-deoxycytidine for demethylation assays.
Results
AE2 promoters were found to be hypermethylated in PBC livers compared to normal and diseased liver specimens. Receiver operating characteristic (ROC) curve analysis showed that minimal CpG-hypermethylation clusters of 3 AE2a-CpG sites and 4 alternate-AE2b2-CpG sites specifically differentiated PBC from normal and diseased controls, with mean methylation rates inversely correlating with respective transcript levels. Additionally, in PBMCs a minimal cluster of 3 hypermethylated AE2a-CpG sites distinguished PBC from controls, and mean methylation rates correlated negatively with AE2a mRNA levels in these immune cells. Alternate AE2b2/AE2b1 promoters in PBMCs were constitutively hypermethylated, in line with absent alternative mRNA expression in diseased and healthy PBMCs. Demethylation assays treating lymphoblastoid-T2 cells with 5-aza-2´-deoxycytidine triggered AE2b2/AE2b1 expression and upregulated AE2a-promoter expression.
Conclusions
Disease-specific hypermethylation of AE2 promoter regions and subsequent downregulation of AE2-gene expression in the liver and PBMCs of patients with PBC might be critically involved in the pathogenesis of this complex disease.
Lay summary
Primary biliary cholangitis (PBC) is a chronic immune-associated cholestatic liver disease with unclear complex/multifactorial etiopathogenesis affecting mostly middle-aged women. Patients with PBC exhibit reduced expression of the AE2/SLC4A2 gene. Herein, we found that AE2 promoter regions are hypermethylated in the liver and peripheral blood mononuclear cells of patients with PBC. This increased methylation is associated with downregulated AE2-gene expression, which might contribute to the pathogenesis of PBC. Therefore, novel epigenetic targets may improve treatment in patients with PBC who respond poorly to current pharmacological therapies.
Keywords: AE2 chloride/bicarbonate exchanger, cholestasis, DNA methylation, epigenetic repression, primary biliary cholangitis/cirrhosis
Graphical abstract
Highlights
-
•
Patients with PBC have higher AE2 CpG methylation in upstream AE2a and/or AE2b2/AE2b1 promoter regions in liver and PBMCs.
-
•
Combined methylation rates of 2 minimal CpG-clusters in the liver and 1 minimal CpG-cluster in PBMCs specifically distinguished PBC from normal and diseased controls.
-
•
Methylation rates of AE2 promoter regions inversely correlated with levels of respective AE2 mRNAs in liver and PBMCs.
-
•
Alternate AE2b2/AE2b1 promoter regions were found to be densely methylated in both normal and diseased PBMC samples.
Introduction
Primary biliary cholangitis (PBC), formerly named primary biliary cirrhosis,1 is a chronic cholestatic liver disease mostly affecting middle-aged women, in which portal mononuclear infiltrates result in progressive damage and destruction of interlobular bile ducts.[2], [3], [4], [5] Thus far, the etiopathogenesis of PBC remains elusive. The disease is regarded as a complex multifactorial disease that may be triggered by environmental factors in individuals with genetic predisposition.6 Genome-wide-association studies (GWAS)[7], [8], [9], [10] and dense fine-mapping association studies11 have related susceptibility to PBC with genetic variations in genes pertinent to immunity like HLA type II, IL12A, IL12RB2 , IL21 and IL21R genes among others. Certainly, the disease is strongly associated with autoimmune phenomena, such as the presence of autoreactive T cells in portal infiltrates and high-titer serum antimitochondrial autoantibodies (AMA) that recognize antigens at the inner mitochondrial membrane. Notwithstanding these features of autoimmunity in PBC, the therapeutic regimes with potent immunosuppressants have shown little efficacy, which is particularly intriguing if compared with the substantial benefit obtained in most patients with early PBC undergoing therapy with ursodeoxycholic acid (UDCA).[12], [13], [14], [15], [16] Since the hydrophilic bile acid UDCA is known to induce bicarbonate-rich choleresis, we have been postulating that primary or secondary abnormalities in the mechanisms responsible for biliary bicarbonate secretion might have a pathogenic role in PBC.[17], [18], [19], [20], [21] Indeed, positron-emission-tomography studies showed that untreated patients with PBC failed to increase biliary bicarbonate in response to secretin, while treatment with UDCA for a few months could reverse this defect.19
In humans, secretin-stimulated biliary bicarbonate secretion is crucial for adequate bile modifications along the biliary tract.[21], [22], [23], [24] Bicarbonate secretion occurs at the apical membrane of bile-duct cells via electroneutral Na+-independent Cl–/HCO3– anion exchange (AE), which can be stimulated by cAMP, the second messenger for secretin signal transduction.22 AE2/SLC4A2 gene-silencing experiments indicated that AE2 is the main carrier for this activity in cholangiocytes.[25], [26], [27] Previously, we reported that bile-duct cells isolated from PBC patients exhibit defective cAMP-stimulated AE activity,20 and that PBC livers have diminished AE2 expression at the luminal membrane of the biliary epithelium.18 Also, we reported that AE2 mRNA levels are decreased in liver biopsies and peripheral blood mononuclear cells (PBMCs) from patients with PBC.17 The notion that diminished AE2 might be involved in PBC pathogenesis received definite support from our findings in Ae2-knockout mice, which exhibit characteristic features resembling PBC.[28], [29], [30]
Conventional genotyping of selected AE2/SLC4A2 tag single-nucleotide polymorphisms (rs2069443, rs2303933, rs2303937, and rs2303941) in Japanese patients with PBC revealed associations with disease susceptibility and/or anti-centromere antibody production.31 But in Caucasian patients, no association between variations in AE2/SLC4A2 and PBC susceptibility has been reported, though single-nucleotide polymorphism analyses across this gene have found 2 variants influencing AMA status.32 Also, a synonymous variation in exon 6 (rs2303932) was related with the progression of the disease in a French population.33 More recent findings provided evidence for upregulation of microRNA 506 in PBC bile-duct cells being involved in the decreased liver expression of AE2 protein through blocking the translation (but not the transcription) of AE2 messages.34 However, the genetic and/or epigenetic factors responsible for the decreased AE2 mRNA expression in PBC17 remain to be fully elucidated. Since hypermethylation of CpG-cytosines in promoter regions is a frequent mechanism for transcriptional inactivation,[35], [36], [37] we explored these possible variations in AE2/SLC4A2 promoter regions. Thus, we analyzed the methylation rates of proximal CpG-cytosines in the widely expressed upstream promoter AE2a and in the tissue-restricted alternate overlapping AE2b2/AE2b1 promoter regions (located within intron 2 of the AE2/SLC4A2 gene),[38], [39] in liver and PBMC samples from patients with PBC and normal and diseased controls. Additionally, we determined the correlation between methylation rates in AE2 promoters and the levels of respective AE2 mRNAs in these samples.
Patients and methods
Human samples and patients
Liver specimens (n = 20) and PBMCs (n = 16) from patients with PBC were obtained at the Clinic University of Navarra (Pamplona) and the Hospital Clinic (Barcelona) before patients started their treatment with UDCA. Liver specimens were frozen samples of either diagnostic liver biopsies or pieces of liver explants from patients transplanted because of advanced PBC. Diagnosis of PBC was based on biochemical cholestasis, AMA positivity (as determined by immunofluorescence), and compatible histology – presymptomatic forms with normal alkaline phosphatase were also considered – (see Table 1 for patient characteristics and biochemical parameters). Pieces of liver explants from patients transplanted because of other liver diseases (OLDs) (n = 24) and PBMCs from additional patients with OLDs (n = 25) were included in the study as non-PBC diseased controls (see Table 2 for patient characteristics and biochemical parameters). The 24 patients with OLDs from whom liver explants were obtained were further split into patients with either moderate or severe cholestasis according to biochemical parameters, as detailed in Table S1. Additionally, normal bordering tissue samples (n = 14) obtained from surgery dissection of metastatic tumors in the liver were included as normal liver controls (NL). PBMCs used as normal controls (n = 16) were anonymized samples obtained from healthy blood-donors/volunteers (HVs). All patients gave written informed consent for the study, which was approved by the institutional Ethics Committees, in compliance with the ethical principles of the Declaration of Helsinki.
Table 1.
Main characteristics of the PBC-patient cohorts⁎
| Liver specimens, n = 20 (mean ± SD) | PBMC specimens, n = 16 (mean ± SD) | |
|---|---|---|
| Sex (female:male) | 18:2 | 15:1 |
| Age (years) | 54.3 ± 13.1 | 52.5 ± 8.0 |
| ALP (IU/L) | 478.6 ± 430.7 | 489.5 ± 308.4 |
| GGT (IU/L) | 238.0 ± 270.1 | 147.8 ± 252.4 |
| ALT (IU/L) | 51.2 ± 34.8 | 46.0 ± 32.7 |
| AST (IU/L) | 45.4 ± 28.7 | 40.6 ± 35.7 |
| Bilirubin (mg/dl) | 1.4 ± 2.2 | 0.8 ± 0.4 |
ALP, serum alkaline phosphatase; ALT, serum alanine aminotransferase; AST, serum aspartate aminotransferase; GGT, serum gamma glutamyltransferase; PBC, primary biliary cholangitis; PBMCs, peripheral blood mononuclear cells.
The most relevant characteristics of the 2 different populations of patients with PBC from whom either liver samples or PBMCs were obtained, are indicated. The mean age of patients from whom normal liver samples were obtained (6 female and 8 male) was 58.2 years (SD: ± 12.9). The PBMC samples employed as normal controls were obtained from healthy volunteers/blood-donors after being completely anonymized.
Table 2.
Main characteristics of patients with OLDs⁎
| Patients with OLDs from whom liver specimens were obtained | |||||||
|---|---|---|---|---|---|---|---|
| Patients (female/male) | Age (years) | ALP (IU/L) | GGT (IU/L) | ALT (IU/L) | AST (IU/L) | Bilirubin (mg/dl) | |
| ALD | 0 / 13 | 56.1 ± 9.4 | 329.3±190.7 | 103.1 ± 99.7 | 41.1 ± 19.0 | 56.2 ± 19.8 | 3.8 ± 1.9 |
| NAFLD | 2 / 2 | 58.9 ± 11.7 | 275.8 ± 92.3 | 199.8 ± 61.2 | 117.3 ± 81.6 | 41.7 ± 14.6 | 0.7 ± 0.4 |
| TxH | 0 / 1 | 70.7 | 1632.0 | 914.0 | 77.0 | 80.0 | 5.4 |
| VHC | 0 / 3 | 59.7 ± 16.6 | 131.0 ± 30.1 | 33.3 ± 21.5 | 44.0 ± 19.3 | 22.7 ± 2.1 | 0.9 ± 0.5 |
| PSC | 0 / 1 | 54.6 | 1598.0 | 309.0 | 52.0 | 48.0 | 5.6 |
| SBC | 1 / 1 | 68.5 ± 4.5 | 574.5±632.9 | 103.0±108.9 | 12.0 ± 5.7 | 16.5 ± 3.5 | 1.0 ± 0.4 |
| Patients with OLDs from whom PBMC specimens were obtained | |||||||
| Patients (female/male) | Age (years) | ALP (IU/L) | GGT (IU/L) | ALT (IU/L) | AST (IU/L) | Bilirubin (mg/dl) | |
| ALD | 1 / 10 | 57.6 ± 5.9 | 202.6 ± 59.2 | 31.4 ± 14.7 | 16.2 ± 10.4 | 28.5 ± 9.7 | 5.6 ± 4.3 |
| NAFLD | 0 / 1 | 46.0 | 109.0 | 38.0 | 70.0 | 28.0 | 1.4 |
| TxH | 1 / 0 | 45.0 | 1321.0 | 815.0 | 71.0 | 76.0 | 5.1 |
| VHC | 5 / 5 | 57.5 ± 13.8 | 124.9 ± 33.6 | 24.7 ± 18.0 | 61.7 ± 45.8 | 44.1 ± 34.0 | 0.6 ± 0.3 |
| PSC | 0 / 1 | 55.0 | 184.0 | 39.0 | 10.0 | 24.0 | 4.5 |
| SBC | 0 / 1 | 59.0 | 133.5 | 27.0 | 9.5 | 22.5 | 4.1 |
ALD, alcohol-related liver disease; ALP, serum alkaline phosphatase; ALT, serum alanine aminotransferase; AST, serum aspartate aminotransferase; GGT, serum gamma glutamyltransferase; NAFLD, non-alcoholic fatty liver disease; OLDs, other liver diseases; PBMCs, peripheral blood mononuclear cells; PSC, primary sclerosing cholangitis; SBC, secondary biliary cirrhosis; TxH, toxic hepatitis; VHC, viral hepatitis C.
The most relevant characteristics (mean ± SD) of patients with OLDs from whom either liver samples or PBMCs were obtained, are indicated. Each OLD population was classified according to the ethology.
Assessment of genomic-DNA methylation and mRNA expression
Genomic DNA (gDNA) and total RNA from liver specimens were extracted with an RNA/DNA Extraction Kit (QIAGEN), while a TRI-Reagent solution (Sigma) was used for both extractions from PBMC samples. DNA methylation rates in proximal AE2 promoter regions (upstream AE2a promoter and alternate AE2b1 and AE2b2 promoters within intron 2)[38], [39] were obtained at 1-base pair resolution by pyrosequencing. Briefly, gDNA aliquots (1 μg) were treated overnight with bisulphite (EpiTect-Bisulphite Kit from QIAGEN) and used as template for PCR amplification with primers that match non-CpG regions within the bisulphite converted promoter sequences (see Table S2). 5’-biotinylated reverse primers allowed for resultant amplicons to be immobilized onto streptavidin-coated beads, followed by denaturation in 0.5 M NaOH, and sequencing of biotinylated reverse strands with respective forward primers (using a PyroMarkTM Q96 pyrosequencer and PyroQ-CpGTM 1.0.9 software; Biotage-QIAGEN).
The levels of mRNAs were assessed by real-time PCR in an iCycler iQ5 (BioRad), using reverse-transcribed total RNA and specific primers for AE2a, AE2b1 and AE2b2, and for GAPDH as normalizing control (Table S3). For calculations, we used the Livak/Schmittgen’s method,40 though modified after estimating an average amplification efficiency of 80%, i.e. 1.8–ΔΔCT.
Lymphoblastoid-T2 cells
Human T2 cells (174xCEM.T2) – hybrid from B and T lymphoblasts – were cultured in RPMI with 25 mM Hepes, 10% fetal bovine serum, and penicillin/streptomycin (Gibco-Invitrogen). Cells (1x105/well) were treated for 48 and 72 h with and without the demethylating agent 5-aza-2´-deoxycytidine (2 μM, Sigma) or the histone-deacetylase inhibitor trichostatin-A (300 nM; Sigma). Nucleic acids were extracted and analyzed as described above for PBMCs.
Statistics
Data are shown as mean ± SD unless otherwise indicated. Statistical analyses were performed by using the GraphPad Prism-5 software (San Diego, CA). Because the number of analyzed samples was usually ~20 for each group, non-parametric tests were regularly employed (even though, according to D’Agostino-Pearson test, there were variables showing normally distributed values). Differences between PBC, OLD and normal control samples were analyzed with Kruskall-Wallis test followed by Mann-Whitney U test to analyze the differences between 2 groups. The Spearman's rank-correlation coefficient rs was also determined when pertinent. Two-tailed p values ≪0.05 were considered statistically significant. Receiver (or Relative) operating characteristic (ROC) curves were constructed and the area under ROC curves (AUC) was calculated to evaluate sensitivity and specificity of multiple CpG-dinucleotide sets within AE2 promoters. The criteria to obtain differentiating CpG-site combinations were ROC curves with p ≪0.05 when PBC was compared with both NL and OLD livers, and ROC curves with p ≫0.05 for comparisons between OLD and normal livers. The cut-off points on the ROC curves at which accuracy of PBC detection was maximal, were selected.
Results
CpG-methylation analysis of AE2 promoters in liver samples
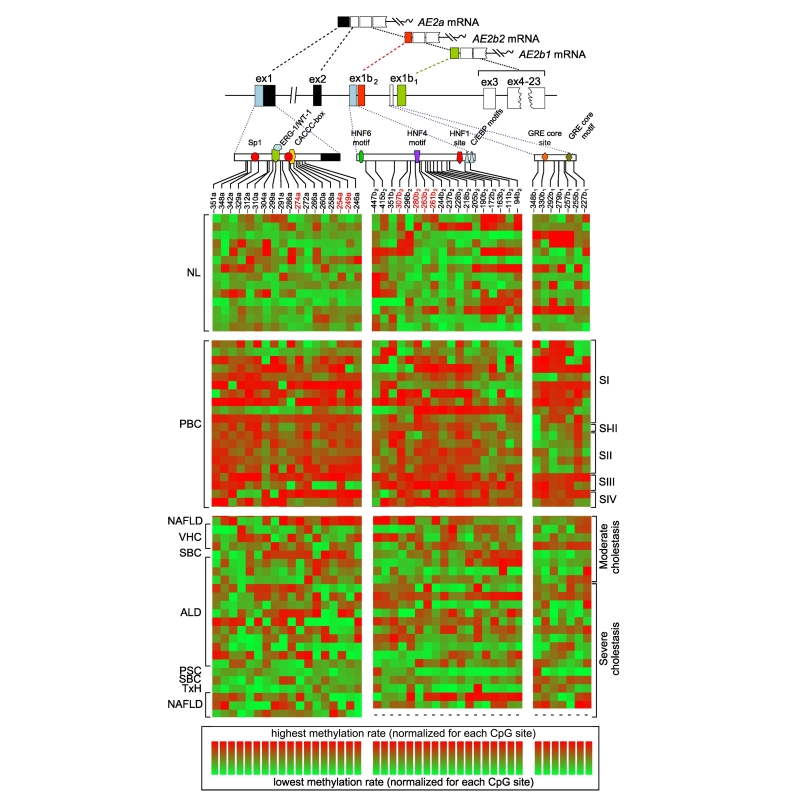
Pyrosequencing of the proximal AE2a promoter region encompassing 18 CpG sites in liver gDNA revealed increased average methylation in PBC livers (24.01 ± 9.60%) compared to NL samples (10.78 ± 4.07%, p ≪0.001) and OLD specimens (13.19 ± 7.31%, p ≪0.005), while no significant differences were observed between OLD and NL samples (see also heat-map representations41 in Fig. 1). Because alternative transcriptional activity is normally prominent in the human liver,39 alternate AE2b2/AE2b1 overlapping promoter regions were also analyzed in liver gDNA. Similarly to our observations for AE2a, pyrosequencing of 18 CpG sites in AE2b2 and 7 CpG sites in AE2b1 regions indicated that average methylation rates in PBC (65.71 ± 11.05% for AE2b1 and 49.14 ± 7.03% for AE2b2) were increased compared to NL samples (56.72 ± 8.99% for AE2b1, p ≪0.05, and 38.55 ± 7.22% for AE2b2, p ≪0.001) and OLD specimens (56.66 ± 10.17% for AE2b1, p ≪0.01, and 40.43 ± 7.38% for AE2b2, p ≪0.001, whereas no significant differences were observed between NL and OLD liver samples (see also heat maps in Fig. 1).
Fig. 1.
Heat maps of CpG-methylation rates for AE2 promoter regions in the liver. The methylation rates of CpG-cytosines within interrogated proximal regions of the upstream AE2a promoter (155-bp long; left panels), and alternate AE2b2 (477-bp; middle panels) and AE2b1 promoters (205-bp; right) were determined through pyrosequencing of bisulphite-converted gDNA from NL specimens (n = 14), PBC-liver samples (n = 20, grouped by disease stages SI-IV), and liver samples from patients with OLDs (n = 24). The upper diagrams show AE2 promoter regions with relevant sites and putative motifs for transcription factors, the initial exons and the transcribed variants. Indicated CpG-dinucleotide positions, i.e. the negative numbers followed by either a, b1 or b2, are referred to cytosine locations upstream to respective +1 position, i.e., A in ATG start codon in exon 2 for AE2a transcript, and in exons 1b1 and 1b2, for AE2b1 and AE2b2 mRNA variants, respectively. Red-labeled numbers indicate hypermethylated CpG sites conforming the PBC-associated methylation signatures of minimal clusters. Maps were obtained with the online Genesis software (http://genome.tugraz.at/serverclient/serverclient_download.shtml, originally designed at Graz University of Technology for microarray data; cf. ref.41). Here, the highest and lowest methylation-rate values – among all values obtained for every CpG site – were normalized in such a way that they ranged from 100% (light red) to 0% (light green) for each site (in accordance with respective color bars below). ALD, alcohol-related liver disease; bp, base pair; gDNA, genomic DNA; NAFLD, non-alcoholic fatty liver disease; NL, normal liver; OLDs, other liver diseases; PBC, primary biliary cholangitis; PSC, primary sclerosing cholangitis; SBC, secondary biliary cirrhosis; TxH, toxic hepatitis; VHC, viral hepatitis C.
AE2 CpG-methylation signatures in PBC livers
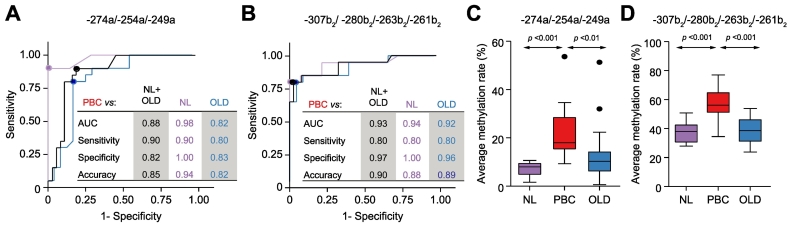
We then searched for minimal clusters of AE2 CpG methylation that could specifically discriminate the PBC condition from both normal and OLD controls. We therefore analyzed average methylation rates of all possible combinations of assessed CpG sites, and performed ROC curve analysis of differentiation between PBC, NL and OLD livers. As illustrated in Fig. 2A-B, 2 minimal CpG clusters, one relative to the AE2a promoter (–274a, –254a, and –249a sites), and the other relative to the AE2b2 alternate promoter (–307b2, –280b2, –263b2, and –261b2 sites), gave highly significant ROC curves, with the highest fraction of PBC samples correctly identified as positive (i.e. highest sensitivity) and with the highest fraction of non-PBC samples correctly identified as negative (i.e. highest specificity). As expected, average methylation rates of these 2 clustered CpG sites were significantly higher in PBC than in normal and diseased controls (Fig. 2C-D). Noticeably, the hypermethylated CpG sites of the AE2b2 minimal cluster (–307b2, –280b2, –263b2, and –261b2 sites) are surrounding an HNF4 motif,39 closely upstream of the HNF1 site,[25], [42] in the AE2b2 alternate promoter (see Fig. 1, upper diagram). Additional ROC curve analysis combining the 2 minimal CpG clusters of AE2a and AE2b2 promoters resulted in the highest AUC (Fig. S1). ROC curve analyses, therefore, allowed us to differentiate the PBC condition from the controls (NL and OLD) with high sensitivity, specificity and accuracy. Altogether, our data support the notion that, in the liver, AE2 promoter regions possess hypermethylation signatures of clustered CpG sites which are able to distinguish PBC from both NL and OLD.
Fig. 2.
PBC-associated methylation signatures of minimal CpG-site clusters differentiating PBC from NL and OLD liver samples. Highly significant ROC curves were obtained for average methylation of (A) a minimal CpG cluster within the AE2a promoter region (–274a, –254a, and –249a sites), and (B) another minimal cluster within the AE2b2 alternate promoter (–307b2, –280b2, –263b2, and –261b2 sites); both ROC curve analyses also included comparisons of PBC versus NL+OLD cohort (non-PBC samples). Analyses always resulted in the highest fraction of PBC samples correctly identified as positive and with the highest fraction of non-PBC samples correctly identified as negative (highest sensitivity and highest specificity, respectively). Box plots to the right (each with minimum value, the first quartile, the median, the third quartile, and the maximum value) graphically depict the comparisons, between PBC and control liver samples, of average methylation rates of PBC-associated minimal CpG clusters in (C) AE2a promoter (–274a, –254a, and –249a sites), and in (D) AE2b2 alternate promoter (–307b2, –280b2, –263b2, and –261b2 sites). Significant p values (≪0.05) were obtained by Kruskall-Wallis test followed by Mann-Whitney U test. NL, normal liver; PBC, primary biliary cholangitis; OLD, other liver disease.
Gender and AE2 promoter hypermethylation
The female preponderance in PBC is remarkable – worldwide, the ratio of affected females to males is as high as 10:1.43 Recently, DNA-methylation sex differences were reported for a series of genes and intergenic regions in the human liver.44 We therefore decided to compare average methylation rates of the PBC-associated CpG clusters in NL from females with those from males, and found no differences, while highly significant differences were observed among females between NL and PBC (Table S4). This indicates gender is not a determinant factor for the hypermethylation changes detected in PBC specimens.
Negative correlations between mRNA levels of AE2 isoforms and methylation rates within respective promoter regions in liver samples
qPCR analysis of the 3 AE2 mRNA isoforms showed their levels were significantly lower in PBC livers than in NL samples (p ≪0.001 for both AE2a and AE2b2 mRNAs and p ≪0.05 for AE2b1 mRNA). Concerning the mRNA levels in our current OLD samples (n = 24), they were markedly lower in the specimen subset from patients with advanced and severe cholestasis (n = 16, Table S1 and Fig. S2), whereas in samples from patients with moderate cholestasis (n = 8, Table S1) the mRNA levels were significantly higher (Fig. S2). In PBC specimens, however, the AE2 mRNA levels were always consistently diminished, regardless of whether patients had severe or moderate cholestasis (Fig. S2).
To assess whether diminished AE2 gene expression was associated with increased promoter methylation we performed correlation analyses between the levels of AE2 mRNA isoforms and average CpG methylation rates. Significant negative correlations were obtained for the cohort including all liver samples, i.e. in the PBC+NL+OLD cohort (Table S5). Correlation coefficients were higher in the cohort including PBC livers and NL samples only (PBC+NL cohort, with no OLDs samples), while analysis of the cohort with just OLD and NL samples (without PBC samples) gave no significant correlations (Table S5). In fact, the methylation rates of AE2 promoter regions in the liver subset of severely cholestatic patients with OLDs were in the range of those in patients with OLDs and moderate cholestasis (Fig. S3), despite the encountered differences in AE2 mRNA levels between the 2 OLD subsets (Fig. S2). These data concur with the view that the diminished AE2 expression observed in the former OLD subset is unrelated to CpG methylation but due to severe cholestasis, while the diminished liver expression of AE2 mRNAs in PBC livers appears to be amply justified by the increased CpG methylation in AE2 promoter regions.
AE2 CpG methylation and AE2 mRNA expression in PBMCs
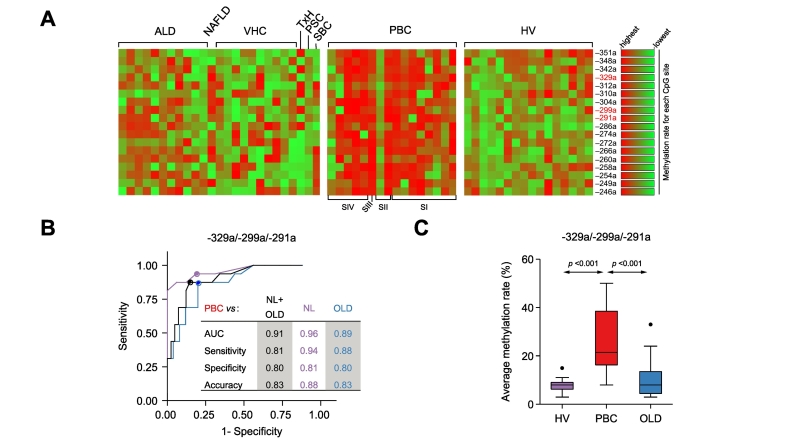
Pyrosequencing analysis of the AE2a promoter in PBMCs showed ≫2-fold increased average methylation in PBC (20.18 ± 8.83%) compared to healthy and diseased controls (HVs 7.16 ± 1.37% and OLDs 9.22 ± 3.04%; p ≪0.01 each; see heat-map representations in Fig. 3A). ROC curve analysis of average methylation rates of all possible combinations of assessed CpG sites revealed a minimal cluster of 3 particular CpG-cytosines (at positions −329a, −299a, and −291a) that specifically differentiated PBC samples from both controls (Fig. 3B). Indeed, the average methylation rate of these combined CpG sites was significantly higher in PBC (26.0 ± 13.22) than in both healthy (7.75 ± 2.93; p ≪0.001) and diseased controls (10.32 ± 7.48; p ≪0.001; Fig. 3C).
Fig. 3.
PBC-associated methylation pattern of AE2a promoter region in PBMCs. (A) Heat map of methylation rates of CpG sites within the interrogated AE2a promoter in PBMCs from HVs (n = 16), patients with PBC (n = 16) and patients with OLDs (n = 25). Maps were obtained and normalized as described in Fig. 1 for the AE2 promoter regions in the liver; here, normalization color bars locate to the right. (B) Highly significant ROC curves obtained for a minimal cluster of methylated CpG sites within AE2a promoter in PBMCs (–329a, –299a, and –291a), comparing PBC versus HV, OLD, and HV+OLD (i.e. non-PBC samples). (C) Comparisons of average methylation rates of the PBC-associated minimal CpG cluster within AE2a promoter in PBMCs (–329a, –299a, and –291a), between PBC and normal and diseased control samples. Significant p values (p ≪0.05) were obtained by Kruskall-Wallis test followed by Mann-Whitney U test. ALD, alcohol-related liver disease; HVs, healthy blood-donors/volunteers; gDNA, genomic DNA; NAFLD, non-alcoholic fatty liver disease; OLDs, other liver diseases; PBC, primary biliary cholangitis; PBMCs, peripheral blood mononuclear cells; PSC, primary sclerosing cholangitis; SBC, secondary biliary cirrhosis; TxH, toxic hepatitis; VHC, viral hepatitis C.
Quantitative-PCR analysis of PBMCs from 12 patients with PBC, from 25 patients with OLDs and from 16 HVs showed consistent expression of AE2a mRNA in all PBMC samples, while alternative AE2b1 and AE2b2 mRNA variants could hardly be detected (not shown). In agreement with our previous findings,17 the levels of AE2a mRNA in PBMCs were significantly diminished in PBC compared to HV and OLD samples (p ≪0.01 each; Fig. S4). Like liver tissue, negative correlations between AE2a mRNA levels and average methylation rates of AE2a promoter were observed in cohorts with PBC and control samples (Table S6).
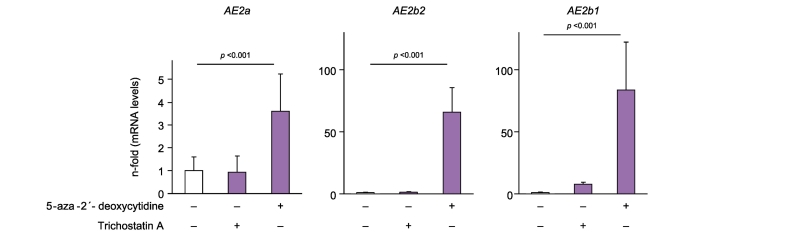
Pyrosequencing of alternate AE2b2 and AE2b1 promoter regions in randomly selected PBMC gDNA samples from individuals with PBC and OLDs, and from HVs revealed consistent methylation (Fig. S5). To investigate whether methylation of alternate AE2b2/AE2b1 promoter regions might hinder the expression of alternative AE2b2 and AE2b1 mRNAs in immune cells we cultured lymphoblastoid T2 cells with and without the demethylating agent 5-aza-2´-deoxycytidine and determined mRNA levels. Baseline, T2 cells were found to express AE2a mRNA but not alternative AE2 variants (Fig. 4), which concurred with the observations in PBMCs. Noticeably, T2 cells treated with 5-aza-2´-deoxycytidine showed substantial expression of alternative AE2b2 and AE2b1 mRNAs (Fig. 4), strongly suggesting that demethylation leads to upregulated transcription from alternate AE2b2/AE2b1 promoters. Moreover, 5-aza-2´-deoxycytidine treatment was followed by a 3.6-fold increase in AE2a mRNA levels (Fig. 4), most probably due to complete release of the AE2a promoter from a baseline status of low-rate methylation (average methylation rate of 8.50 ± 8.25%).
Fig. 4.
CpG methylation is involved in the dormant expression of alternative AE2b1 and AE2b2 mRNAs in human lymphoblastoid T2 cells. Baseline, the overall methylation rates of AE2 promoter regions in T2 cells are the following: 8.50 ± 8.25% for the upstream AE2a region and 61.83 ± 26.11% and 94.14 ± 6.31% for the alternate AE2b2 and AE2b1 regions (respectively). The levels of alternative AE2b2 and AE2b1 mRNAs as well as those of the complete AE2a transcript were determined by real-time PCR in cultured T2 cells treated for 48 h with either 5-aza-2´-deoxycytidine, trichostatin-A, or just vehicle as control. Values (normalized for GAPDH mRNA) are given as fold expression relative to values of the control with vehicle alone. Significant p values refer to comparisons versus the control with vehicle. Significant p values (p ≪0.05) were obtained by Kruskall-Wallis test followed by Mann-Whitney U test.
Finally, we tested a possible role of histone acetylation-deacetylation for AE2 transcription and treated T2 cells with the HDAC inhibitor trichostatin-A during 48 and 72 h. In contrast to the effects observed with 5-aza-2´-deoxycytidine, no significant changes in the levels of mRNAs were found with trichostatin-A (see Fig. 4 depicting the results at 48 h).
Discussion
Here we report that significant epigenetic modifications of the AE2 gene occur in the liver and PBMCs of patients with PBC, which could explain the reduced gene expression in these patients. Thus, we observed an increased CpG-cytosine methylation of AE2a, AE2b1 and AE2b2 proximal promoters in PBC livers compared to normal and diseased controls. Hypermethylation affects more intensely particular CpG sites in each promoter region. ROC curve analysis found 2 minimal clusters of hypermethylated CpG-cytosines (1 comprised of 3 CpG-cytosines in the upstream AE2a promoter, and another comprised of 4 CpG sites in the alternate AE2b2 promoter, see Fig. 2), which could specifically differentiate PBC from both normal and OLD samples. Moreover, average methylation rates of combined CpG-cytosines were found to inversely correlate with the levels of AE2 mRNAs driven by respective promoter regions in the liver.
Analogous data and correlations were observed in PBMCs but referred only to the upstream AE2a promoter and AE2a transcript. ROC curve analyses also found a minimal cluster composed by 3 CpG-cytosines in the AE2a promoter, the average methylation of which could specifically differentiate PBMC samples of patients with PBC from normal and OLD peripheral immune cells (Fig. 3). On the other hand, alternate AE2b2/AE2b1 promoter regions were ascertained to be silent not only in PBC but also in normal and diseased PBMCs. These regions show constitutive increased CpG-cytosine methylation that presumably prevents the expression of alternative AE2 transcripts. This view is strongly supported by our findings in lymphoblastoid T2 cells, in which dormant expression of AE2b2 and AE2b1 mRNAs could be activated when T2 cells were treated with the demethylating agent 5-aza-2´-deoxycytidine. Interestingly, such a treatment also increased the expression of AE2a mRNA, revealing that further release from basal low-rate methylation in the AE2a proximal promoter might result in amplified transcriptional activity. Treatment with trichostatin-A, however, had no effect on AE2 promoter activities in T2 cells, which suggests that histone deacetylation is poorly involved in dormant alternative expression of AE2 in these immune cells.
Altogether, our findings indicate that hypermethylation of the AE2 promoter regions might be an important mechanism leading to decreased expression of AE2 mRNAs in the liver and PBMCs of patients with PBC. It may be speculated that methylation of CpG-cytosines located in the vicinity of particular motifs might preclude the recruitment of DNA binding proteins and cause transcriptional inactivation.[35], [36] Interestingly, the minimal cluster of CpG-cytosines in the AE2a promoter that are hypermethylated in PBC livers spans near the equivalent minimal cluster detected in PBMCs from patients with PBC; for instance, most AE2a-CpG sites with increased methylation in liver and PBMC samples from patients with PBC locate close to Sp1 and ERG-1/WT1 motifs (Fig. 1, Fig. 3). Consequently, the clusters detected in the AE2a promoter region (1 in liver samples and another in PBMCs from patients with PBC) can affect similar motifs in the promoter. It might also be speculated that methylated cytosines serve as docking sites for methyl-CpG-binding domain (MBD) proteins and repress transcription indirectly via recruitment of corepressors and chromatin modification.[35], [36]
Transcription from alternate AE2b2 and AE2b1 overlapping promoter regions is quite restricted to the liver and kidney in humans, when compared to rodents.39 Most probably, liver restriction is connected to motifs for liver-enriched and related transcription factors like HNF1, CBP, and glucocorticoid receptor encountered in those overlapping promoter regions in the human gene.[25], [42] Of notice, most AE2b2/AE2b1-CpG sites characteristically hypermethylated in the liver of PBC patients locate in the neighborhood of sites and motifs for those liver-related transcription factors.39 In fact, the minimal cluster of hypermethylated CpG-cytosines in the AE2b2 alternate promoter which discriminates PBC livers from normal liver samples and OLD samples spans a region encompassing an HNF1 site[25], [42] and an upstream HNF4 motif.39
In the liver, AE2 is involved in biliary bicarbonate secretion.[26], [27] This exchanger protein is therefore directly responsible for creating the "bicarbonate umbrella" along the biliary tree that protects cholangiocytes from the proapoptotic effects of bile salts by maintaining them deprotonated.45 In a context of reduced liver AE2 expression and AE2 deficiency, there may occur defective bicarbonate umbrella and subsequent entry of protonated bile salts into cholangiocytes (concisely and nicely reviewed in ref.12). Entry of highly hydrophobic bile salts can promote reactive oxygen species (ROS) production, and lead to enhanced inflammatory cytokine and chemokine responses.46 ROS production might also contribute to further decreasing AE2 expression.46 On the other hand, AE2 knockdown in vivo was shown to result in increased ROS production in the liver.46 Additionally, AE2 knockdown experiments in cholangiocytes led to intracellular accumulation of bicarbonate and increased expression and activity of soluble adenylyl cyclase (sAC).47 This bicarbonate sensor was found to sensitize cholangiocytes to bile salt-induced apoptosis through the intrinsic apoptotic pathway.47 Interestingly, cholangiocytes were previously reported to translocate immunologically intact PDC-E2 to apoptotic bodies and create an apotope,48 that may favor the development of AMAs. The triad of bile-duct cell derived apotopes, macrophages from patients with PBC, and AMAs were also found to trigger an intense production of proinflammatory cytokines.49 More recently, cultured human cholangiocytes treated with proinflammatory cytokines typically overexpressed in PBC livers (such as IL8, IL12, IL17, IL18, and TNF-α) were shown to enhance the expression of miR-506, that induced PBC-like features in these bile-duct cells and promoted immune activation.50 Noticeably, bile-duct cells of PBC livers were described to upregulate miR-506, which can inhibit translation of AE2 transcripts and contribute to further decreasing the expression of AE2 protein.34
Previously reported experiments in animal models showing that highly hydrophobic bile salts might decrease AE2 mRNA expression through ROS production46 seemingly have a correlate in the subset of our current diseased liver control specimens from severely cholestatic patients (Fig. S2). In this subset of diseased control samples, we found markedly low levels of AE2 mRNAs despite AE2 promoter methylation being comparable to that in normal-liver control samples (as well as to those in diseased control samples from patients with OLDs and moderate cholestasis; Fig. S3). In PBC livers, however, promoter methylation was significantly increased (Fig. S3) and AE2 mRNA levels were consistently diminished (Fig. S2), regardless of whether patients had severe or moderate cholestasis. Altogether, these data support the notion that diminished AE2 gene expression in PBC livers primarily results from increased AE2 CpG methylation, whereas in OLD specimens from severely cholestatic patients, the diminished liver expression of AE2 gene occurs as a direct consequence of the elevated levels of hydrophobic bile salts, unrelated to CpG methylation. Our earlier findings in Ae2-knockout mice,28 as well as the aforementioned AE2 knockdown experiments (both in vivo46 and in bile-duct cells47), clearly show that a primary failure in AE2 gene expression may result in detrimental cascades in the liver.
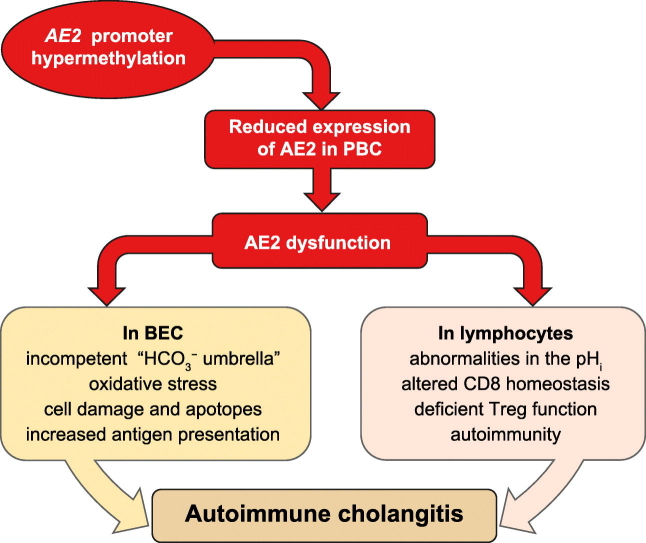
In PBMCs, AE2 is involved in intracellular pH regulation and immunological homeostasis.[24], [29], [30] Thus AE2 promoter hypermethylation and deficient AE2 mRNA expression in patients with PBC may contribute to the immune dysregulation PBC patients typically have. Indeed the particular predisposition to activation of immune cells against bile-duct cells in PBC resembles the dysfunctions of T cells observed in our Ae2-knockout animal model, in which they interact in the liver with highly immunogenic AE2-deficient bile-duct cells.[28], [29], [30] In contrast to PBC, no autoimmune cholangitis is expected to develop in severely cholestatic patients with OLDs despite dramatically decreased AE2 mRNA levels in the liver, since PBMCs do not appear to be equally affected by the severe cholestasis. Thus, the idiosyncratic conjunction of anomalies putatively streaming from AE2 hypermethylation in both the liver and PBMCs in patients with PBC may constitute a crucial 2-arms prerequisite for the development of autoimmune cholangitis (Fig. S6), and provide relevant clues to unravel the enigmatic pathogenesis of the disease.
A series of epigenetic alterations on the X chromosome and autosomal chromosomes were previously reported in PBC.[51], [52], [53] Some genes appear hypermethylated while other genes are with decreased DNA methylation, indicating that hypermethylation is not a widespread phenomenon in PBC. Therefore, the increased methylation of the AE2 gene currently observed in our study in PBC samples might be viewed as disease specific in the context of additional epigenetic modifications. Disclosing the cause(s) of such modifications in PBC should be a priority for the related studies in the near future. Certainly, PBC is a multifactorial liver disease that may ensue from highly complex interactions between genetic and environmental factors, as stressed by GWAS data and more conventional genetic and epidemiological studies.[6], [54], [55], [56] Environmental exposures may result in epigenetic modifications, particularly DNA methylation.57 Smoking, for instance, which has been associated with high risk of PBC,[56], [58] was also related with increased DNA-methyltransferase expression.59 Therefore, DNA methylation affecting AE2 and additional genes, may constitute a link between genetic and environmental risk factors in PBC pathogenesis.
In summary, AE2 deficiency in patients with PBC may render cholangiocytes more immunogenic and susceptible to autoimmune attack, whereas an equivalent defect in PBMCs may alter immunological homeostasis.[29], [30] Additional identified (and not yet identified) genetic and epigenetic modifications in genes pertinent to immunity may further boost a pivotal contribution of AE2 deficiency, and lead to immune dysregulation and autoimmunity (Fig. S6).
Financial support
This work was supported by an agreement between FIMA and the “UTE for CIMA Project”, the Spanish Ministry of Education and Science SAF2009-11538 (JFM) and the Carlos III Institute of Health-CIBEREHD Program (JFM, JP, AP).
Declaration of Competing Interest
The authors declare no conflicts of interest that pertain to this work.
Please refer to the accompanying ICMJE disclosure forms for further details.
Authors’ contributions
JFM made the conception and design of the study. FA, JP and AP also contributed to the study design. FA, IH, ES and SM collected the data. FA, JFM and AP performed the statistical analysis. JFM, FA, JP and AP contributed to the interpretation of the data. FA, JFM and AP drafted and edited the manuscript. JFM, JP and AP were responsible for funding acquisition.
Acknowledgments
The authors thank Ms Virginia Villar and Silvia Ruiz-Gaspà for their valuable help with sample collection, and Dr. Axel R. Concepción for his constructive comments.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.jhepr.2019.05.006.
Supplementary data
Supplementary material 1
Supplementary material 2
Supplementary material 3
References
- 1.Beuers U, Gershwin ME, Gish RG, Invernizzi P, Jones DE, Lindor K. Changing nomenclature for PBC: From 'cirrhosis' to 'cholangitis'. J Hepatol. 2015;63:1285–1287. doi: 10.1016/j.jhep.2015.06.031. [DOI] [PubMed] [Google Scholar]
- 2.Trivedi PJ, Cullen S. Etiopathogenesis of primary biliary cirrhosis: an overview of recent developments. Hepatol Int. 2013;7:28–47. doi: 10.1007/s12072-012-9362-7. [DOI] [PubMed] [Google Scholar]
- 3.Hohenester S, Oude-Elferink RP, Beuers U. Primary biliary cirrhosis. Semin Immunopathol. 2009;31:283–307. doi: 10.1007/s00281-009-0164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poupon R. Primary biliary cirrhosis: a 2010 update. J Hepatol. 2010;52:745–758. doi: 10.1016/j.jhep.2009.11.027. [DOI] [PubMed] [Google Scholar]
- 5.Kumagi T, Heathcote EJ. Primary biliary cirrhosis. Orphanet J Rare Dis. 2008;3 doi: 10.1186/1750-1172-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gershwin ME, Mackay IR. The causes of primary biliary cirrhosis: Convenient and inconvenient truths. Hepatology. 2008;47:737–745. doi: 10.1002/hep.22042. [DOI] [PubMed] [Google Scholar]
- 7.Hirschfield GM, Liu X, Xu C, Lu Y, Xie G, Lu Y. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. N Engl J Med. 2009;360:2544–2555. doi: 10.1056/NEJMoa0810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirschfield GM, Liu X, Han Y, Gorlov IP, Lu Y, Xu C. Variants at IRF5-TNPO3, 17q12-21 and MMEL1 are associated with primary biliary cirrhosis. Nat Genet. 2010;42:655–657. doi: 10.1038/ng.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mells GF, Floyd JAB, Morley KI, Cordell HJ, Franklin CS, Shin SY. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet. 2011;43:329–332. doi: 10.1038/ng.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu F, Tang R, Zuo X, Shi X, Wei Y, Zheng X. A genome-wide association study identifies six novel risk loci for primary biliary cholangitis. Nat Commun. 2017;14:14821–14828. doi: 10.1038/ncomms14828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu JZ, Almarri MA, Gaffney DJ, Mells GF, Jostins L, Cordell HJ. Dense fine-mapping study identifies new susceptibility loci for primary biliary cirrhosis. Nat Genet. 2012;44:1137–1141. doi: 10.1038/ng.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molinaro A, Marschall HU. Why doesn't primary biliary cholangitis respond to immunosuppressive medications? Curr Hepatology Rep. 2017;16:119–123. doi: 10.1007/s11901-017-0345-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corpechot C, Carrat F, Bahr A, Chretien Y, Poupon RE, Poupon R. The effect of ursodeoxycholic acid therapy on the natural course of primary biliary cirrhosis. Gastroenterology. 2005;128:297–303. doi: 10.1053/j.gastro.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Parés A, Caballería L, Rodés J. Excellent long-term survival in patients with primary biliary cirrhosis and biochemical response to ursodeoxycholic acid. Gastroenterology. 2006;130:715–720. doi: 10.1053/j.gastro.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 15.Lindor K. Ursodeoxycholic acid for the treatment of primary biliary cirrhosis. N Engl J Med. 2007;357:1524–1529. doi: 10.1056/NEJMct074694. [DOI] [PubMed] [Google Scholar]
- 16.Kuiper EM, Hansen BE, de Vries RA, den Ouden-Muller JW, van Ditzhuijsen TJ, Haagsma EB. Improved prognosis of patients with primary biliary cirrhosis that have a biochemical response to ursodeoxycholic acid. Gastroenterology. 2009;136:1281–1287. doi: 10.1053/j.gastro.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 17.Prieto J, Qian C, García N, Díez J, Medina JF. Abnormal expression of anion exchanger genes in primary biliary cirrhosis. Gastroenterology. 1993;105:572–578. doi: 10.1016/0016-5085(93)90735-u. [DOI] [PubMed] [Google Scholar]
- 18.Medina JF, Martínez-Ansó E, Vázquez JJ, Prieto J. Decreased anion exchanger 2 immunoreactivity in the liver of patients with primary biliary cirrhosis. Hepatology. 1997;25:12–17. doi: 10.1002/hep.510250104. [DOI] [PubMed] [Google Scholar]
- 19.Prieto J, García N, Martí-Climent JM, Penuelas I, Richter JA, Medina JF. Assessment of biliary bicarbonate secretion in humans by positron emission tomography. Gastroenterology. 1999;117:167–172. doi: 10.1016/s0016-5085(99)70564-0. [DOI] [PubMed] [Google Scholar]
- 20.Melero S, Spirlì C, Zsembery A, Medina JF, Joplin RE, Duner E. Defective regulation of cholangiocyte Cl–/HCO3– and Na+/H+ exchanger activities in primary biliary cirrhosis. Hepatology. 2002;35:1513–1521. doi: 10.1053/jhep.2002.33634. [DOI] [PubMed] [Google Scholar]
- 21.Medina JF. Role of the anion exchanger 2 in the pathogenesis and treatment of primary biliary cirrhosis. Dig Dis. 2011;29:103–112. doi: 10.1159/000324144. [DOI] [PubMed] [Google Scholar]
- 22.Banales JM, Prieto J, Medina JF. Cholangiocyte anion exchange and biliary bicarbonate excretion. World J Gastroenterol. 2006;12:3496–3511. doi: 10.3748/wjg.v12.i22.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beuers U, Hohenester S, de Buy Wenniger LJ, Kremer AE, Jansen PL, Elferink RP. The biliary HCO3– umbrella: A unifying hypothesis on pathogenetic and therapeutic aspects of fibrosing cholangiopathies. Hepatology. 2010;52:1334–1340. doi: 10.1002/hep.23810. [DOI] [PubMed] [Google Scholar]
- 24.Concepcion AR, López M, Ardura-Fabregat A, Medina JF. Role of AE2 for pHi regulation in biliary epithelial cells. Front Physiol. 2014;4:413. doi: 10.3389/fphys.2013.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arenas F, Hervías I, Úriz M, Joplin R, Prieto J, Medina JF. Combination of ursodeoxycholic acid and glucocorticoids upregulates the AE2 alternate promoter in human liver cells. J Clin Invest. 2008;118:695–709. doi: 10.1172/JCI33156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banales JM, Arenas F, Rodríguez-Ortigosa CM, Sáez E, Uriarte I, Doctor RB. Bicarbonate-rich choleresis induced by secretin in normal rat is taurocholate-dependent and involves AE2 anion exchanger. Hepatology. 2006;43:266–275. doi: 10.1002/hep.21042. [DOI] [PubMed] [Google Scholar]
- 27.Úriz M, Sáez E, Prieto J, Medina JF, Banales JM. Ursodeoxycholic acid is conjugated with taurine to promote secretin-stimulated biliary hydrocholeresis in the normal rat. PLoS One. 2011;6 doi: 10.1371/journal.pone.0028717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salas JT, Banales JM, Sarvide S, Recalde S, Ferrer A, Uriarte I. Ae2a,b-deficient mice develop antimitochondrial antibodies and other features resembling primary biliary cirrhosis. Gastroenterology. 2008;134:1482–1493. doi: 10.1053/j.gastro.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 29.Concepcion AR, Salas JT, Sarvide S, Sáez E, Ferrer A, López M. Anion exchanger 2 is critical for CD8+ T cells to maintain pHi homeostasis and modulate immune responses. Eur J Immunol. 2014;44:1341–1351. doi: 10.1002/eji.201344218. [DOI] [PubMed] [Google Scholar]
- 30.Concepcion AR, Salas JT, Sáez E, Sarvide S, Ferrer A, Portu A. CD8+ T cells undergo activation and programmed death-1 repression in the liver of aged Ae2a,b−/− mice favoring autoimmune cholangitis. Oncotarget. 2015;6:28588–28606. doi: 10.18632/oncotarget.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aiba Y, Nakamura M, Joshita S, Inamine T, Komori A, Yoshizawa K. Genetic polymorphisms in CTLA4 and SLC4A2 are differentially associated with the pathogenesis of primary biliary cirrhosis in Japanese patients. J Gastroenterol. 2011;46:1203–1212. doi: 10.1007/s00535-011-0417-7. [DOI] [PubMed] [Google Scholar]
- 32.Juran BD, Atkinson EJ, Larson JJ, Schlicht EM, Lazaridis KN. Common genetic variation and haplotypes of the anion exchanger SLC4A2 in primary biliary cirrhosis. Am J Gastroenterol. 2009;104:1406–1411. doi: 10.1038/ajg.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poupon R, Ping C, Chretien Y, Corpechot C, Chazouilleres O, Simon T. Genetic factors of susceptibility and of severity in primary biliary cirrhosis. J Hepatol. 2008;49:1038–1045. doi: 10.1016/j.jhep.2008.07.027. [DOI] [PubMed] [Google Scholar]
- 34.Banales JM, Saez E, Uriz M, Sarvide S, Urribarri AD, Splinter P. Up-regulation of microRNA 506 leads to decreased Cl–/HCO3− anion exchanger 2 expression in biliary epithelium of patients with primary biliary cirrhosis. Hepatology. 2012;56:687–697. doi: 10.1002/hep.25691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clouaire T, Stancheva I. Methyl-CpG binding proteins: specialized transcriptional repressors or structural components of chromatin? Cell Mol Life Sci. 2008;65:1509–1522. doi: 10.1007/s00018-008-7324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 37.Carvalho ATP, Gouveia L, Kanna CR, Wärmländer SKT, Platts JA, Kamerlin SCL. Understanding the structural and dynamic consequences of DNA epigenetic modifications: Computational insights into cytosine methylation and hydroxymethylation. Epigenetics. 2014;9:1604–1612. doi: 10.4161/15592294.2014.988043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Medina JF, Acín A, Prieto J. Molecular cloning and characterization of the human AE2 anion exchanger (SLC4A2) gene. Genomics. 1997;39:74–85. doi: 10.1006/geno.1996.4467. [DOI] [PubMed] [Google Scholar]
- 39.Medina JF, Lecanda J, Acín A, Ciesielczyk P, Prieto J. Tissue-specific N-terminal isoforms from overlapping alternate promoters of the human AE2 anion exchanger gene. Biochem Biophys Res Commun. 2000;267:228–235. doi: 10.1006/bbrc.1999.1951. [DOI] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 41.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 42.Malumbres R, Lecanda J, Melero S, Ciesielczyk P, Prieto J, Medina JF. HNF1α upregulates the human AE2 anion exchanger gene (SLC4A2) from an alternate promoter. Biochem Biophys Res Commun. 2003;311:233–240. doi: 10.1016/j.bbrc.2003.09.200. [DOI] [PubMed] [Google Scholar]
- 43.Lleo A, Battezzati PM, Selmi C, Gershwin ME, Podda M. Is autoimmunity a matter of sex? Autoimmun Rev. 2008;7:626–630. doi: 10.1016/j.autrev.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 44.García-Calzón S, Perfilyev A, de Mello VD, Pihlajamäk J, Ling C. Sex differences in the methylome and transcriptome of the human liver and circulating HDL-cholesterol levels. Clin Endocrinol Metab. 2018;103:4395–4408. doi: 10.1210/jc.2018-00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hohenester S, de Buy Wenniger LM, Paulusma CC, van Vliet SJ, Jefferson DM, Oude Elferink RP. A biliary HCO3– umbrella constitutes a protective mechanism against bile acid-induced injury in human cholangiocytes. Hepatology. 2012;55:173–183. doi: 10.1002/hep.24691. [DOI] [PubMed] [Google Scholar]
- 46.Hisamoto S, Shimoda S, Harada K, Iwasaka S, Onohara S, Chong Y. Hydrophobic bile acids suppress expression of AE2 in biliary epithelial cells and induce bile duct inflammation in primary biliary cholangitis. J Autoimmun. 2016;75:150–160. doi: 10.1016/j.jaut.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 47.Chang JC, Go S, de Waart DR, Munoz-Garrido P, Beuers U, Paulusma CC. Soluble adenylyl cyclase regulates bile salt-induced apoptosis in human cholangiocytes. Hepatology. 2016;64:522–534. doi: 10.1002/hep.28550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lleo A, Selmi C, Invernizzi P, Podda M, Coppel RL, Mackay IR. Apotopes and the biliary specificity of primary biliary cirrhosis. Hepatology. 2009;49:871–879. doi: 10.1002/hep.22736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lleo A, Bowlus CL, Yang GX, Invernizzi P, Podda M, Van de Water J. Biliary apotopes and anti-mitochondrial antibodies activate innate immune responses in primary biliary cirrhosis. Hepatology. 2010;52:987–998. doi: 10.1002/hep.23783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erice O, Munoz-Garrido P, Vaquero J, Perugorria MJ, Fernandez-Barrena MG, Sáez E. MicroRNA-506 promotes primary biliary cholangitis-like features in cholangiocytes and immune activation. Hepatology. 2018;67:1420–1440. doi: 10.1002/hep.29533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lleo A, Liao J, Invernizzi P, Zhao M, Bernuzzi F, Ma L. Immunoglobulin M levels inversely correlate with CD40 ligand promoter methylation in patients with primary biliary cirrhosis. Hepatology. 2012;55:153–160. doi: 10.1002/hep.24630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Selmi C, Cavaciocchi F, Lleo A, Cheroni C, De Francesco R, Lombardi SA. Genome-wide analysis of DNA methylation, copy number variation, and gene expression in monozygotic twins discordant for primary biliary cirrhosis. Front Immunol. 2014;5:121–129. doi: 10.3389/fimmu.2014.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lleo A, Zhang W, Zhao M, Tan Y, Bernuzzi F, Zhu B. DNA methylation profiling of the X chromosome reveals an aberrant demethylation on CXCR3 promoter in primary biliary cirrhosis. Clin Epigenetics. 2015;7:61. doi: 10.1186/s13148-015-0098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Selmi C, Meda F, Kasangian A, Invernizzi P, Tian Z, Lian Z. Experimental evidence on the immunopathogenesis of primary biliary cirrhosis. Cell Mol Immunol. 2010;7:1–10. doi: 10.1038/cmi.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones DE. Pathogenesis of primary biliary cirrhosis. Gut. 2007;56:1615–1624. doi: 10.1136/gut.2007.122150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prince MI, Ducker SJ, James OF. Case-control studies of risk factors for primary biliary cirrhosis in two United Kingdom populations. Gut. 2010;59:508–512. doi: 10.1136/gut.2009.184218. [DOI] [PubMed] [Google Scholar]
- 57.Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol Metab. 2010;21:214–222. doi: 10.1016/j.tem.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gershwin ME, Selmi C, Worman HJ, Gold EB, Watnik M, Utts J. Risk factors and comorbidities in primary biliary cirrhosis: a controlled interview-based study of 1032 patients. Hepatology. 2005;42:1194–1202. doi: 10.1002/hep.20907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hammons GJ, Yan Y, Lopatina NG, Jin B, Wise C, Blann EB. Increased expression of hepatic DNA methyltransferase in smokers. Cell Biol Toxicol. 1999;15:389–394. doi: 10.1023/a:1007658000971. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2
Supplementary material 3