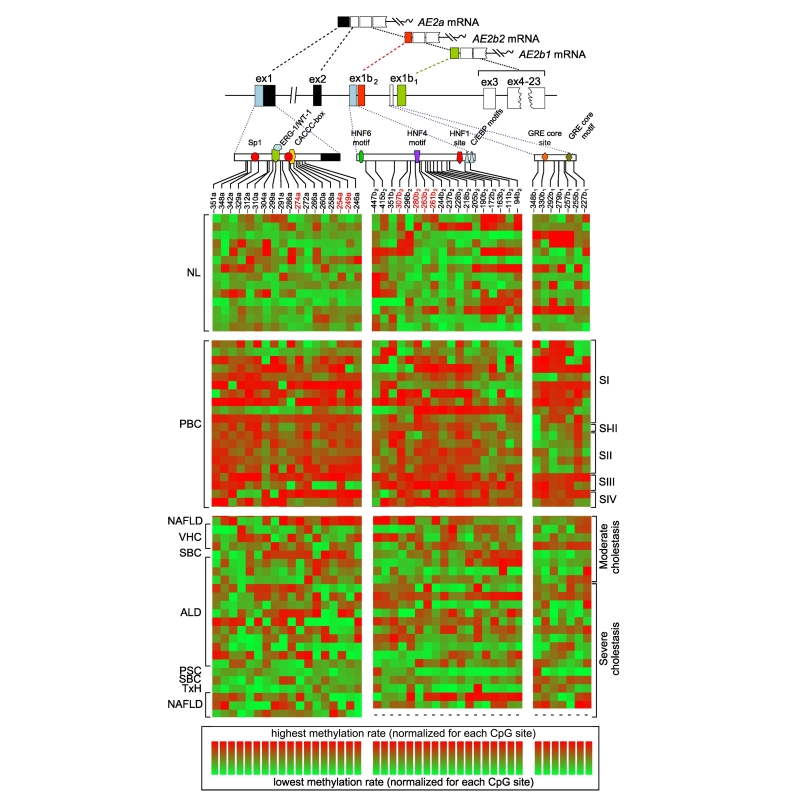
Fig. 1.
Heat maps of CpG-methylation rates for AE2 promoter regions in the liver. The methylation rates of CpG-cytosines within interrogated proximal regions of the upstream AE2a promoter (155-bp long; left panels), and alternate AE2b2 (477-bp; middle panels) and AE2b1 promoters (205-bp; right) were determined through pyrosequencing of bisulphite-converted gDNA from NL specimens (n = 14), PBC-liver samples (n = 20, grouped by disease stages SI-IV), and liver samples from patients with OLDs (n = 24). The upper diagrams show AE2 promoter regions with relevant sites and putative motifs for transcription factors, the initial exons and the transcribed variants. Indicated CpG-dinucleotide positions, i.e. the negative numbers followed by either a, b1 or b2, are referred to cytosine locations upstream to respective +1 position, i.e., A in ATG start codon in exon 2 for AE2a transcript, and in exons 1b1 and 1b2, for AE2b1 and AE2b2 mRNA variants, respectively. Red-labeled numbers indicate hypermethylated CpG sites conforming the PBC-associated methylation signatures of minimal clusters. Maps were obtained with the online Genesis software (http://genome.tugraz.at/serverclient/serverclient_download.shtml, originally designed at Graz University of Technology for microarray data; cf. ref.41). Here, the highest and lowest methylation-rate values – among all values obtained for every CpG site – were normalized in such a way that they ranged from 100% (light red) to 0% (light green) for each site (in accordance with respective color bars below). ALD, alcohol-related liver disease; bp, base pair; gDNA, genomic DNA; NAFLD, non-alcoholic fatty liver disease; NL, normal liver; OLDs, other liver diseases; PBC, primary biliary cholangitis; PSC, primary sclerosing cholangitis; SBC, secondary biliary cirrhosis; TxH, toxic hepatitis; VHC, viral hepatitis C.