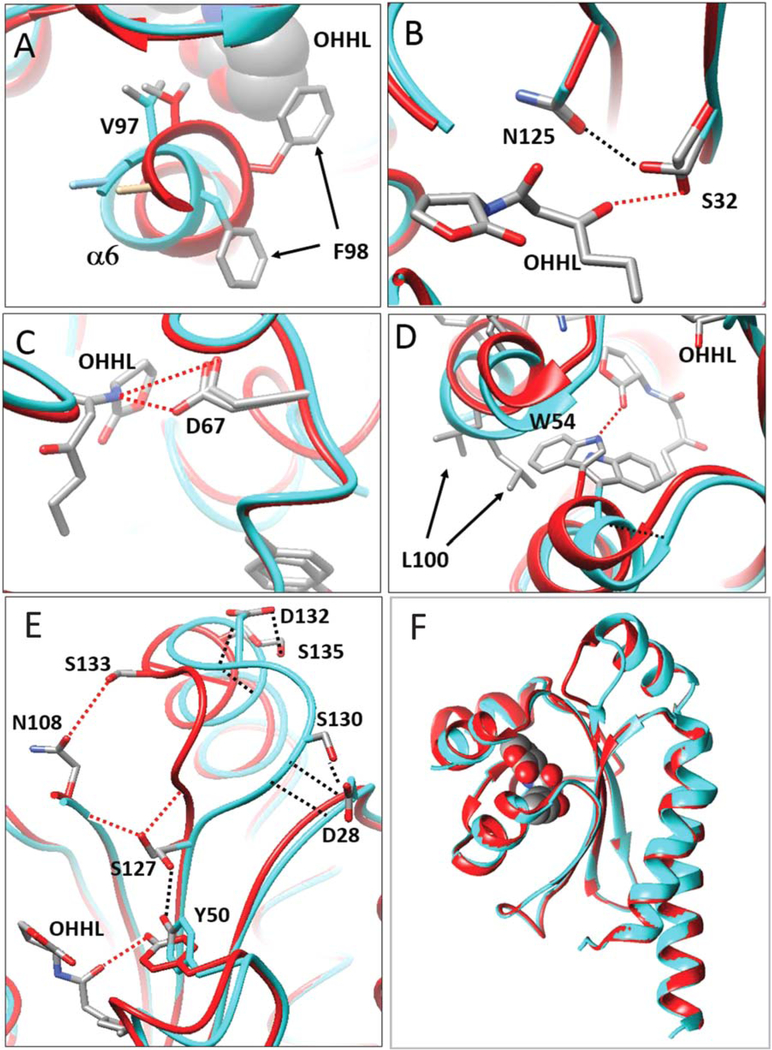
FIGURE 4.
OHHL changes the conformation of residues at the binding site. Apoprotein is depicted in cyan, while the ligand-bound form is depicted in red. (A) OHHL causes Phe98 to swing inward, causing a distortion of helix a6. In this conformation, Val97 is drawn close to OHHL, contributing to the hydrophobic environment. (B) Ser32 makes a hydrogen bond with OHHL, while in the apoprotein, Ser32 swivels 90° to bond with Asn125. (C) Asp67 forms a hydrogen bond with OHHL, and makes little or no movement in response to OHHL. (D) Trp54 extends well into the binding pocket of the apoprotein, while OHHL causes Trp54 to swivel approximately 120°, avoiding a steric clash, and allowing a hydrogen bond between it and OHHL. This outward movement necessitates a movement of the nearby residue Leu100, which protrudes into solvent. (E) Binding of OHHL causes a movement of a loop between β4 and α7. In the apoprotein, Tyr50 bonds with Ser127, and the loop (cyan) is pulled to the right by bonds between Asp28 and Gly129-Ser130, and by bonds between Ser135 and Asp131-Asp132 (black lines). OHHL bonds with Tyr50, releasing the bond between Try50 and Ser127. Ser127 then can swivel and bond to the backbone of Asn108 and Gly129 (red lines). This pulls the loop (red) toward Asn108. This conformation is further stabilized by bonds between Ser133 and Asn108, and between Asn128 and Asp131 (not shown). (F) The loop described in part E is the only extensive conformational change in the protein induced by OHHL. All other tertiary structures can be closely superimposed [Color figure can be viewed at wileyonlinelibrary.com]