Figure 4.
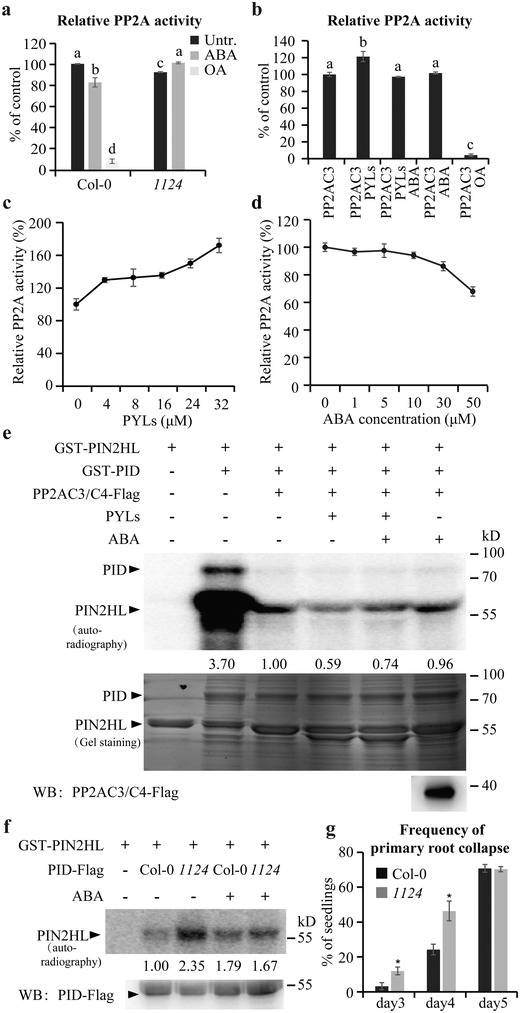
PYLs regulate PP2A activity and thus PID‐mediated PIN phosphorylation. a) An in vivo PP2A activity assay. The total protein used for the assay was extracted from 10‐d‐old seedlings treated or not with 30 × 10−6 m ABA for 4 h. The activity of PP2A in untreated wild type was set to 100%. b) An in vitro enzyme activity assay showing the effect of PYLs and/or ABA on the PP2A activity of PP2AC3. The concentrations of PYLs and ABA were 8 × 10−6 and 5 × 10−6 m, respectively. The phosphatase activity of PP2AC3 alone was set to 100%. PP2A activity was always measured after addition of 1 × 10−3 m EDTA to inhibit the activity of PP2C (a,b). OA was used as a phosphatase inhibitor (a,b). Error bars represent ± SE of three replicates. Means with different letters (a,b) are significantly different at P < 0.05 (Fisher LSD test). c) PYLs enhancing the phosphatase activity of PP2AC3 in a dosage‐dependent manner in vitro. The concentrations of PYLs protein were 0, 4 × 10−6, 8 × 10−6, 16 × 10−6, 24 × 10−6, and 32 × 10−6 m, respectively. The phosphatase activity of PP2AC3 alone was set to 100%. d) ABA dosage‐dependent reduction of the PYLs‐increased PP2AC3 activity in vitro. 4 × 10−6 m PYLs protein was incubated with different concentrations of ABA before mixing with PP2AC3 for phosphatase activity assay. The phosphatase activity of the sample without ABA treatment was set to 100%. Error bars represent ± SE of three replicates (c,d). e) An in vitro phosphorylation assay reconstituting reversible PIN2HL phosphorylation by ABA, PYLs, and/or PP2ACs. The PP2AC3/C4 expressed in Arabidopsis protoplasts, and recombinant PYLs proteins (PYR1, PYL1, PYL2, PYL4), PIN2HL, and PID expressed in E. coli were used (b–e). The concentrations of PYLs and ABA used for the phosphorylation assay were 30 × 10−6 and 50 × 10−6 m, respectively. Arrowheads mark positions of the different proteins. f) PID‐mediated PIN phosphorylation affected by ABA as well as its receptor PYLs. Protoplasts from wild type and 1124 mutant expressing PID‐Flag were treated or not with 10 × 10−6 m ABA for 4 h. PID‐Flag was immunoprecipitated and coincubated with purified recombinant GST‐PIN2HL protein from E. coli in a phosphorylation reaction. The proteins were finally separated by SDS‐PAGE. Arrowheads mark positions of the different proteins. Numbers under lanes indicate relative band intensities normalized to the loading controls (e,f). + and − indicate incubated with or without substrate, extracts, or ABA treatment, respectively (e,f). g) Quantification of 35S::PID‐mediated root collapse in 3‐, 4‐, and 5‐d‐old seedlings (n ≥ 120 roots). The collapse was significantly promoted by PYLs mutation. Error bars represent ± SE of three replicates. (∗) P < 0.05 (Student's t‐test). Three independent experiments were performed with similar results. Representative images are shown.
