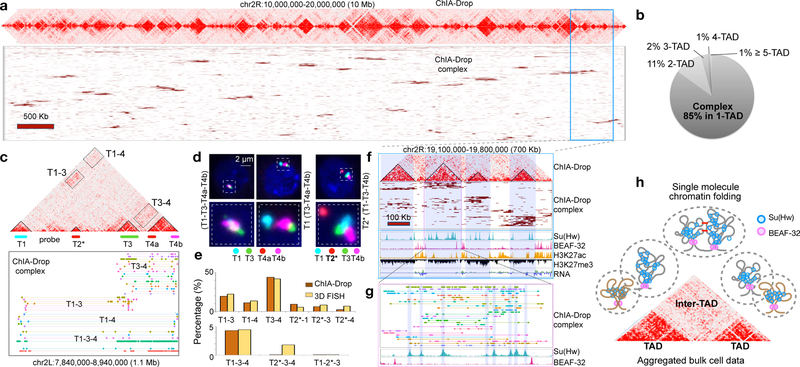
Figure 2. Multiplex chromatin interactions from ChIA-Drop.
a. ChIA-Drop data shown as “linear multi-fragment views” of complexes with ≥10 fragments. Each row is a complex and columns are 50 Kb bins.
b. Proportion of chromatin complexes in single TAD (1-TAD) or multiple TADs (2- to 5-TAD).
c. ChIA-Drop data containing 4 TADs. Fluorescent DNA probes T1-T4b were designed.
d. FISH images. Left, 4 test probes colocalized (T1-T3-T4a-T4b) in 17 of 404 nuclei; middle, 3 test probes colocalized (T3-T4a-T4b) in 177 of 404 nuclei; right, the control T2* probe far from the other probes in 221 of 221 nuclei. Data are combined from two replicates.
e. Percentage of ChIA-Drop complexes involving a given combination of TADs and the corresponding FISH colocalization (Top, 2-TADs; Bottom, 3-TADs). 150 nuclei were analyzed with a 0.28 μm “colocalization” cutoff.
f. A 700 kb region in chr2R shows ChIA-Drop complexes binned at 10 kb, along with RNA-Seq and ChIP-Seq of Su(Hw), BEAF-32, H3K27ac, and H3K27me3.
g. Further zoom of chromatin complexes (n=29) with their fragments (n=328) (top), along with Su(Hw) and BEAF-32 profiles (bottom).
h. Proposed single-molecule model of chromatin folding in topological domains. BEAF-32 defines TAD boundaries. Within domains, chromatin is folded randomly and associated with Su(Hw). Nearby domains likely have random or specific inter-domain contacts (red bars).