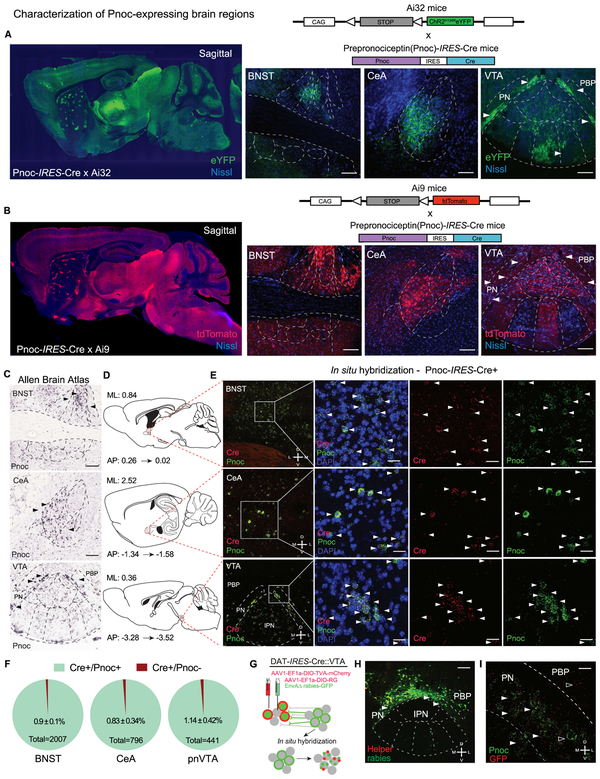
Figure 1: Anatomical Identification of Endogenous Pnoc-expressing VTA Inputs.
(A-B) Generation of the Pnoc-CretdTomato and Pnoc-CreChR2/eYFP mouse lines from the cross between the Pnoc-Cre × Ai9-tdTomato or Pnoc-Cre × Ai32-ChR2/eYFP. Sagittal images of Pnoc labeling in Pnoc-Cre × Ai9 or Ai32 with coronal images depicting bed nucleus of stria terminalis (BNST), central amygdala (CeA), and paranigral ventral tegmental area (pnVTA). Images show tdTomato (red) or ChR2/eYFP (green) and Nissl (blue) staining. Scale bars are 100 μm.
(C) Representative coronal images for Pnoc in situ hybridization from Allen Brain Institute of the BNST, CeA, VTA/IPN regions. All scale bars are 100 μm.
(D) Sagittal atlas images depicting the location of BNST, CeA, and VTA regions used for quantification.
(E) Pnoc and Cre expression patterns in Pnoc-Cre+ mice via in situ hybridization of Pnoc (green), Cre (red), and DAPI (blue) for coronal images of BNST, CeA, and pnVTA regions corresponding to panel D. Scale bars are 25 μm.
(F) Quantification of Pnoc expression within Cre-expressing cells in the BNST, CeA, and pnVTA (n=4, 4 mice, 3 slices each)
(G) Schematic depicting in situ hybridization for GFP (red) and Pnoc (green) mRNA following recombinant AAV helper viral and rabies viral injections into the VTA of DAT-Cre mice.
(H) Coronal image depicting cre-dependent helper (red) and monosynaptic rabies (green) viral expression in the VTA / IPN regions of DAT-Cre mice. PBP- parabrachial pigmented nucleus, PN – paranigral VTA. Scale bars are 200 μm.
(I) Coronal image depicting fluorescent in situ hybridization (FISH) of Pnoc (green) and GFP (red) colocalization in the pnVTA of DAT-Cre mice following recombinant AAV helper viral and rabies viral injections into the VTA. Scale bars are 25 μm.