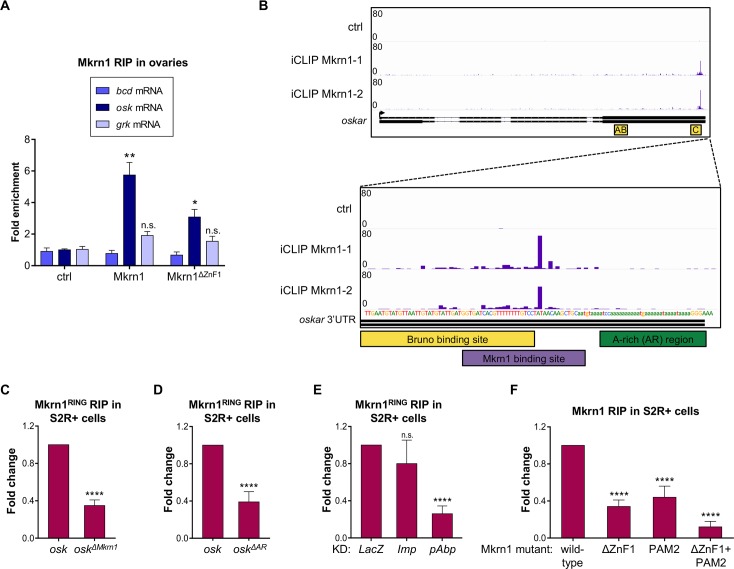
Fig 6. Mkrn1 associates specifically with the 3’ UTR of osk mRNA.
(A) RIP experiment. Either FLAG-tagged Mkrn1 or Mkrn1ΔZnF1 was overexpressed in Mkrn1N ovaries using nos>GAL4 driver. Enrichment of different transcripts was analyzed by RT-qPCR using primers that bind to the respective 3’ UTRs. Fold enrichment is presented relative to the control (nos>GAL4 driver alone, ctrl). Error bars depict SEM, n = 3. Multiple t-test was used to analyze significant changes compared to control RIP. (B) iCLIP results from S2R+ cells showing specific binding of Mkrn1 to osk in a region of the 3’ UTR that partially overlaps with the BRE-C site (yellow). The peaks (purple) indicate crosslinking events of Mkrn1 to osk. Data of two technical replicates for FLAG-Mkrn1 is shown. The same experiment performed with FLAG-GFP (ctrl) did not show specific peaks. (C) RIP experiments of FLAG-Mkrn1RING in S2R+ cells. Enrichment of luciferase-osk-3’UTR transcript was analyzed by RT-qPCR compared to IP experiments with FLAG-GFP. Mkrn1RING Binding to osk 3’ UTR is compromised when introducing a deletion of the Mkrn1 binding site (oskΔMkrn1, deletion of nucleotides 955–978 of osk 3’ UTR). Error bars depict SEM, n = 4. (D) Binding of Mkrn1RING to luciferase-osk-3’UTR reporter is reduced in S2R+ cells when using a deletion of the A-rich region of osk 3’ UTR (oskΔAR, deletion of nucleotides 987–1019 of osk 3’ UTR). Fold change illustrates the difference of pulled-down oskΔAR reporter compared to IP with wild-type osk 3’ UTR. The RIP experiments were normalized to FLAG-tagged GFP. Error bars depict SEM, n = 4. (E) RIP experiments were performed in either control cells or upon depletion of Imp or pAbp. Enrichment was calculated compared to FLAG-GFP. The relative change in Mkrn1RING binding to luciferase-osk-3’UTR reporter in knockdown cells compared to LacZ-depleted cells is depicted. Depletion of pAbp compromises binding of Mkrn1. Error bars depict SEM, n = 3. (F) RIP experiments showing that FLAG-Mkrn1 binding to luciferase-osk-3’UTR is dependent on the ZnF1 domain as well as on the PAM2 motif. Enrichment was analyzed using RT-qPCR and the relative change in binding compared to RIP of FLAG-Mkrn1 is illustrated. RIP experiments were performed in S2R+ cells using FLAG-GFP as control. Error bars depict SEM, n ≥ 3.