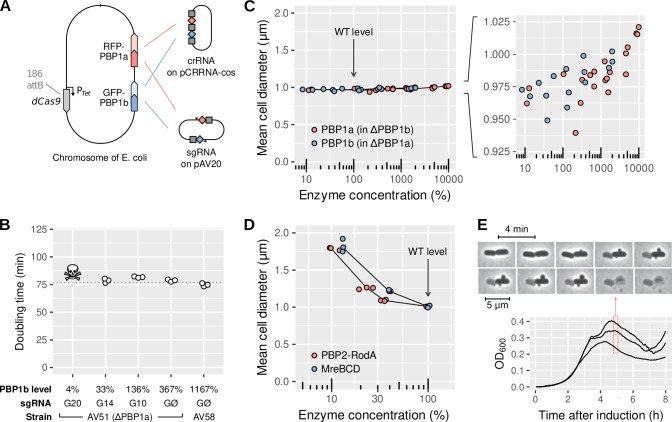
Figure 1. aPBPs have no role in maintaining rod-like cell shape.
(A) Sketch of the strain AV44 (LC69 mrcB::gfp-mrcB, mrcA::rfp-mrcA) with tunable levels of RFP-PBP1a and GFP-PBP1b. CRISPR guides are expressed either as crRNA (top) or as sgRNA (bottom), see also Figure 1—figure supplement 1. (B) Doubling time of AV51 (AV44 ΔPBP1a)/pAV20 as a function of PBP1b level, in minimal medium with glucose and casamino-acids at 30°C. sgRNA are expressed from pAV20 as annotated. AV58 is AV51 Para-GFP-PBP1b for over-expression. Skull logo: not viable. (C) Effect of aPBP concentration on cell diameter. Points indicate the median diameter within each population. Green: AV51/pCRRNAcos with crRNA G20, G14, G10 and GØ, or AV58 (over-expression). Red: AV50 (AV44 ΔPBP1b)/pCRRNAcos with crRNA R20, R18, R11 and RØ. AV63 is AV50 HK022::Para-RFP-PBP1a for over-expression. Levels were determined based on fluorescence and normalized with respect to WT according to DIA. (D) Effect of the concentration of Rod-complex proteins on cell diameter. Green: AV88 (LC69 MreB-GFP)/pAV20 with sgRNA G14, G10 or GØ. Red: AV08 (LC69 RFP-PBP2)/pAV20 with crRNA R20, R18, R11 or RØ. (E) Growth curve of AV44/pAV20 with PBP1ab repressed to lethal level (sgRNA G20-R20), and cell morphology during lysis. Individual points are biological replicates. OD: optical density. WT: wild-type.