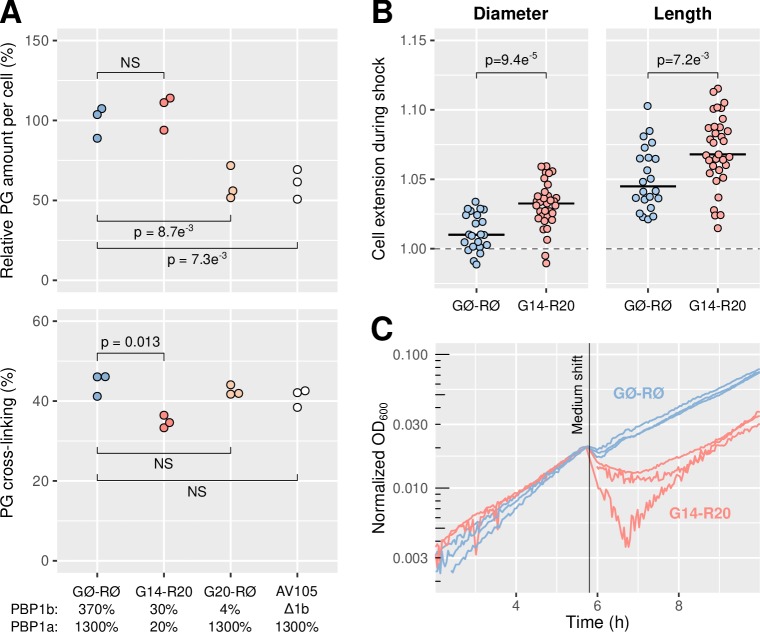
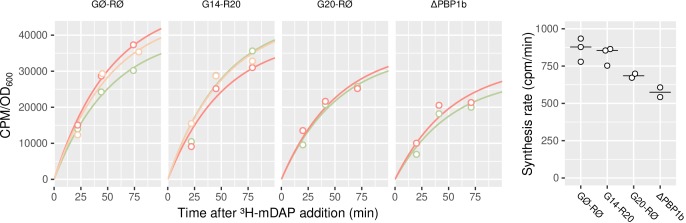
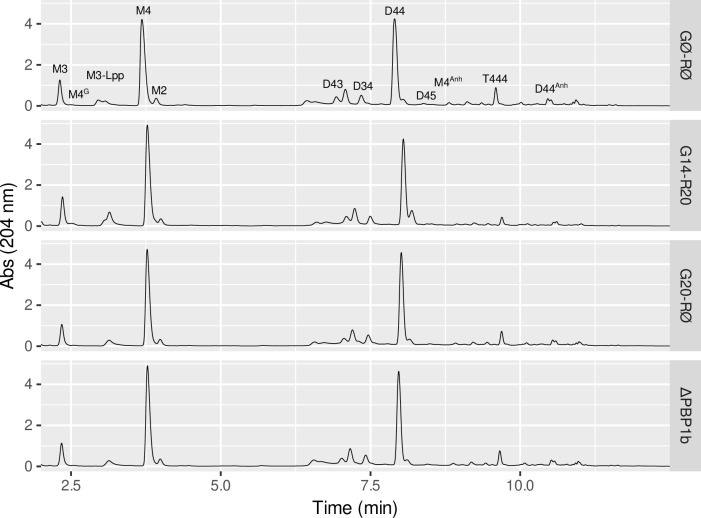
Figure 2. Repression of PBP1ab reduces mechanical stiffness while maintaining a high rate of peptidoglycan insertion.
(A) Top: Steady-state amount of peptidoglycan per cell, measured in AV84 (AV44 ΔLysA)/pAV20 and AV105 (AV44 ΔPBP1b ΔLysA) as annotated. Bottom: Fraction of the peptidoglycan that is cross-linked in the same conditions. p-Values correspond to a two-sided t-test. NS: not significant. (B) Extension of the cells’ short axis (left) and long axis (right) after a 1 osm/L NaCl downshock, in AV93 (AV44 ΔMscLS)/pAV20 with sgRNA GØ-RØ or G14-R20. A value of one corresponds to no extension. Horizontal lines represent the medians. p-Values correspond to a two-sided permutation test. (C) Growth curves before and after a 1 osm/L osmotic downshock, in AV93/pAV20 with sgRNA GØ-RØ or G14-R20. The curves are scaled so all curves have the same OD at the moment of the shock. OD: optical density.