Abstract
Vector‐borne diseases often originate from wildlife and can spill over into the human population. One of the most important determinants of vector‐borne disease transmission is the host preference of mosquitoes. Mosquitoes with a specialised host preference are guided by body odours to find their hosts in addition to carbon dioxide. Little is known about the role of mosquito host preference in the spillover of pathogenic agents from humans towards animals and vice versa. In the Republic of Congo, the attraction of mosquitoes to primate host odours was determined, as well as their possible role as malaria vectors, using odour‐baited traps mimicking the potential hosts of mosquitoes. Most of the mosquito species caught showed a generalistic host preference. Anopheles obscurus was the most abundant Anopheles mosquito, with a generalistic host preference observed from the olfactory response and the detection of various Plasmodium parasites. Interestingly, Culex decens showed a much higher attraction towards chimpanzee odours than to human or cow odours. Human Plasmodium parasites were observed in both human and chimpanzee blood, although not in the Anopheles mosquitoes that were collected. Understanding the role of mosquito host preference for cross‐species parasite transmission provides information that will help to determine the risk of spillover of vector‐borne diseases.
Keywords: Anopheles, Plasmodium, chimpanzee, Congo, bridge vectors, mosquito host preference, transmission dynamics
The majority of the mosquito species caught showed a generalistic host preference.
Anopheles obscurus was the most abundant Anopheles mosquito, with a generalistic host preference, whereas Culex decens showed a much higher attraction towards chimpanzee odours than to human or cow odours
Human Plasmodium parasites were observed in both human and chimpanzee blood, although not in the Anopheles mosquitoes that were collected.

Introduction
Many of the most deadly human diseases, including malaria, Zika virus, dengue virus and chikungunya virus, are transmitted by mosquitoes (Takken & Knols 1999; Harrington et al., 2001; Lambrechts et al., 2010; Fauci and Morens, 2016). In addition to transmitting diseases between human hosts, mosquitoes also facilitate the spillover of zoonotic pathogens into human populations (Jones et al., 2008). As the closest relatives of Homo sapiens, great apes are of interest as potential reservoirs of zoonotic vector‐borne pathogens themselves, and mosquitoes may act as a bridge vector between great ape reservoirs of such pathogens and humans. There is, however, a fundamental gap in knowledge concerning the transmission dynamics of vector‐borne diseases between great apes and humans, as well as the potential zoonotic threat that they pose to humans.
Studies of Plasmodium parasites in African great apes revealed that the malaria parasites Plasmodium falciparum and Plasmodium vivax, which together account for the majority of global malaria cases (WHO World Malaria Report 2018), both originated from parasites that infect African great apes (Liu et al., 2010, 2014; Loy et al., 2018a). Indeed, African great apes are infected with at least 13 Plasmodium species, which are further subdivided into the Laverania and Plasmodium subgenera (Liu et al., 2010, 2014, 2016; Loy et al., 2016,2017). The Laverania parasites tend to be host restricted, whereas Plasmodium parasites appear to have a more promiscuous host tropism.
Although several studies have failed to detect evidence of ape Plasmodium parasites in modern African humans ( Sundararaman et al., 2013; Délicat‐Loembet et al., 2015; Loy et al., 2018b), it is not clear whether this is representive of biological barriers to infection or a lack of exposure to ape parasites. Interestingly, studies of sanctuary apes show that the Laverania host‐species restriction observed in the wild can be broken when chimpanzees and gorillas are housed in the same sanctuary (Ngoubangoye et al., 2016), suggesting that perhaps ecological factors (such as the frequency of exposure to infectious mosquitoes) impact cross‐species transmission.
Although two studies have identified some of the Anopheles species that transmit ape Plasmodium parasites (Paupy et al., 2013; Makanga et al., 2016), little is known about the behaviour of mosquitoes that could serve as bridge vectors between ape and human hosts. One understudied area is the biting behaviour and host species preferences of the mosquitoes that are found in the forest near Plasmodium‐infected apes. Biting behaviour is largely dependent on the mosquito's host preference, which in turn is influenced by the body odour profile of the vertebrate hosts (Takken & Verhulst 2017). Some mosquito species have a specialised attraction towards humans (anthropophilic species), whereas others have a more opportunistic host preference (generalistic species) (Busula et al., 2015). This preference is mediated by differences in volatile compounds produced by different host species (Busula et al., 2017) and similarities in the odour profile of host species could mediate the transmission of pathogens between these species (Verhulst et al., 2012; Verhulst et al., 2018).
In the present study, experiments were performed at the Tchimpounga Chimpanzee Rehabilitation Centre in the Republic of Congo, where chimpanzees were previously reported to harbour ape Plasmodium parasites (Pacheco et al., 2013), aiming to examine the feeding behaviour and host choice of local mosquito species. Mosquito traps were baited with different host volatiles to assess the behaviour of mosquitoes towards different host odours. The present study also aimed to characterize the transmission dynamics of ape Plasmodium parasites by screening chimpanzees and mosquitoes for Plasmodium species.
Materials and methods
Location
Mosquito and chimpanzee blood samples were collected at the Jane Goodall Institute (JGI) Tchimpounga Chimpanzee Rehabilitation Centre (TC) in the Tchimpounga National Reserve, Republic of Congo (4°24′S, 11°48′E). Chimpanzee blood and faecal samples were collected by the veterinary staff of TC from October to November 2015 and October to January 2016, respectively. Mosquito samples were collected from October 2015 to January 2016 at three small islands within the TC that act as natural enclosures. The islands are located in the Kouilou river and house a dormitory where chimpanzees sleep during the night. The chimpanzees can roam free over the islands during the day. Sample collection was approved by the Ministry of Forest Economy and Sustainable Development of the Republic of Congo under permit No. 071. Chimpanzee blood samples were exported by the JGI TC under the Republic of Congo Cites export Permit No. 007008 and 010 (26 February 2016) and imported by the University of Pennsylvania under U.S. Cites Import Permit No. 15US7151B/9 and PHS Import Permit No. 2015‐10‐089. Mosquitoes were imported by the University of Pennsylvania under U.S. PHS Import Permit No. 2016‐02‐084.
Mosquito trapping
Using odour‐baited mosquito traps, the host preference and species composition of a population of mosquitoes can be determined (Qiu et al., 2007). Two different odour‐baited traps were used in the present study: the BG‐Sentinel trap (BioGents GmbH, Regensburg, Germany) and the Suna trap ( BioGents GmbH, Regensburg, Germany). Carbon dioxide(CO2) was supplemented to each trap because it is a general long‐distance mosquito activator and attractant (Schmied et al., 2008; Smallegange et al., 2010).
To test for the most appropriate mosquito trap for trapping Anopheles mosquitoes in the Tchimpounga National Reserve, the BG‐Sentinel trap was compared with the Suna trap. The traps were baited with a standardized five‐component odour blend (Menger et al., 2014; Pombi et al., 2014) and CO2 to attract mosquitoes (Smallegange et al., 2010). Three traps of each type were placed along a transect 30 m apart with similar spatial conditions and rotated using a complete randomized design resulting in six trapping nights.
Carbon dioxide was produced using sugar‐fermenting yeast as an organic source of CO2. Cane sugar and molasses were both used as sugar source. Carbon dioxide was produced using 125 g of cane sugar or molasses, 9 g of yeast and 1 L of water in a 1.5‐L bottle (Mweresa et al., 2014). Additionally, to increase CO2 production, CO2 was produced using 250 g of cane sugar or molasses with 17 g of yeast and 2 L of water in a 5‐L bottle (Mweresa et al., 2014). Water from the Kouilou River was used during field experiments. The CO2 was released using 70 cm of silicon tubing inserted in the cap of the bottle and connected to the mosquito trap.
Host odours from chimpanzees, cows and humans were collected using nylon socks (20 DEN, 100X polyamide; HEMA, De Bilt, The Netherlands). Cow and chimpanzee odours were collected by rubbing a nylon sock on the arm (for chimpanzees) or upper leg (for cows) of the animal for 30 s. Human odour was collected from local male individuals wearing a nylon sock overnight. To minimize the influence of anthropogenic compounds, these individuals did not use deodorant or any perfumed substances the day before wearing a nylon sock overnight. Animal and human odours were collected from three different individuals. The socks were cut into three pieces and one piece of each individual were combined. To determine the most suitable Anopheles trap, a synthetic blend mimicking human odour containing ammonia, (S)‐lactic acid, tetradecanoic acid, 3‐methyl‐1‐butanol and butan‐1‐amine (Menger et al. 2014) was released from the two different trap models, Suna and BG‐sentinel. A clean nylon sock was used as a control. All nylon socks were handled with clean latex gloves and stored in glass jars.
The mosquito traps were operated from 17.00 h to 06.30 h. Suna traps were hung with the trap entry located 30 cm above ground level (Hiscox et al., 2014), whereas the BG‐Sentinel trap was placed on the ground (Schmied et al., 2008). Latex gloves were worn when handling the traps to avoid contamination with odours and the traps were cleaned with 70% ethanol after each trapping night (Busula et al., 2015).
Odour preference of different mosquito species
To assess the attractiveness of mosquitoes to natural host odours, five odour‐baited Suna traps were baited with five different odour treatments consisting of (a) sock with no odour and no CO2; (b) sock with no odour with CO2; (c) sock with cow odour and CO2; (d) sock with chimpanzee odour and CO2; and (v) sock with human odour and CO2. Each trap was set at least 30 m apart and odour baits randomized in a 5 × 5 Latin square experimental design that was repeated seven times. Location one was 4 m in front of a chimpanzee dormitory housing 23 chimpanzees; location two was behind a house where three to four caregivers were constantly present, as well as close (15 m) to the chimpanzee dormitory; location three was near a small settlement of wooden houses with humans; location four was at the edge of a forest, 10 m from wooden houses occupied by humans; and location five was in the forest with no humans and chimpanzees within a 60 m range.
Chimpanzee blood sampling
Blood samples were collected from 84 chimpanzees (Pan troglodytes) living at the Tchimpounga Chimpanzee Rehabilitation Centre. Samples were collected in ethylenediamine tetraacetic acid collection tubes after routine veterinary examination. Density gradient centrifugation was used to separate red blood cells (RBC). To concentrate RBCs, 2–3 mL of whole blood was diluted in phosphate‐buffered saline (1:1 v/v) and gently placed on one volume of Lymphoprep (Axis‐Shield, Oslo, Norway). The mixture was then centrifuged at approximately 700 rpm for 35 min. After removal of the plasma, the RBC were mixed with RNAlater® (1:1 v/v) (Thermo Fisher, Waltham, MA, U.S.A.) in a 15‐mL tube. The plasma was mixed with 1:1 (v/v) RNAlater® in a 15‐mL tube. Blood samples were stored at −20 °C before shipment, shipped at ambient temperature and stored at −80 °C upon arrival. DNA was extracted from chimpanzee blood samples using the DNeasy Blood and Tissue kit (Qiagen, Valencia, CA, U.S.A.).
Mosquito identification
Mosquitoes collected from the traps were killed using ethyl acetate (non‐acetonic nail polish, Kruidvat, The Netherlands) and morphologically identified to genus level using taxonomic keys (Highton, 1983). Anopheles mosquitoes were identified to species level (Gillies and Coetzee, 1987). After morphological identification, Anopheles mosquitoes were stored in 200 μL tubes with RNAlater® (Ambion, Austin, TX, U.S.A.) and stored at −20 °C. Non‐Anopheles mosquitoes were placed in 1.5‐mL Eppendorf tubes with silica beads (Sigma‐Aldrich, St Louis, MO, U.S.A.) and stored at −20 °C. Anopheles mosquitoes identified as part of a species complex were further identified by polymerase chain reaction (PCR) amplification of the Cytochrome oxidase subunit II (COII) gene as described previously by Ndo et al. (2010). Positive PCR products for COII were purified and sequenced using Sanger sequencing. Blood meals from culicine mosquitoes (n = 244) were preserved on filter paper cards (Whatman FTA cards; GE Heathcare, Chicago, IL, U.S.A.). The blood meals were spread onto FTA cards by pressing the blood out of the abdomen using a sterile pipet tip. The FTA cards with blood spots were dried for 1–2 h at room temperature and stored in a CloneSaver pouch (GE Healthcare) together with silica gel (Sigma‐Aldrich) for 1–3 months. Mosquitoes and FTA cards were shipped at ambient temperature and stored at −80 °C before nucleic acid extraction.
DNA extraction
DNA was extracted from Anopheles mosquitoes using the DNeasy 96 Blood & Tissue Kit (Qiagen) in accordance with the manufacturer's instructions for DNA purification from insects. Briefly, mosquitoes were placed in 1.5‐mL Eppendorf tubes containing 180 μL of Buffer ATL (Qiagen) and 20 μL of proteinase K (Qiagen) and crushed using a 1‐mL pipette tip or mortar. Samples were incubated at 56 °C overnight in an incubator. DNA was further extracted in accordance with the manufacturer's instructions. DNA was extracted from 1900 individual mosquitoes and the remainder (3500 mosquitoes) were pooled (five mosquitoes per pool) to reduce the cost of DNA extraction. After DNA extraction, 50 μL of the DNA extract was treated with a One Step™ PCR inhibitor removal kit (Zymo Research, Irvine, CA, U.S.A.) before PCR amplification.
DNA was extracted from CloneSaver FTA filter paper cards (GE Healthcare) using the Allprep DNA/RNA mini kit (Qiagen). Briefly, two or three disks of the dried blood spots were punched out using a Harris 3‐mm micro‐puncher (GE Healthcare). Two or three discs were mixed in a 1,5‐mL Eppendorf tube with 350 μL of RLT buffer (Qiagen) containing 1% β‐mercaptoethanol and incubated for 1 h at 37 °C with shaking (1000 rpm). Afterwards, DNA/RNA was extracted in accordance with the manufacturer's instructions.
Plasmodium parasite screening
All blood and mosquito samples were screened for presence of Plasmodium DNA using pan‐Plasmodium primers targeting the mitochondrial cytochrome b gene (cytB) (956 bp) by nested PCR as described previously (Liu et al., 2010). Amplicons were sequenced directly without interim cloning using Sanger sequencing technology. All obtained mosquito positive sequences were aligned with GenBank reference sequences (see Supporting information, Table S1) using clustalw, version 2.1 (http://www.clustal.org) in geneious, version 11 (https://www.geneious.com). jmodeltest, version 2.1.7 (Darriba et al., 2012) was used to select for best evolutionary model. Maximum likelihood phylogenies with bootstrap support (1000 replicates) were estimated using phyml, version 3 (http://www.atgc-montpellier.fr/phyml) and GTR + G + I as evolutionary models (Guindon et al., 2010).
Nucleotide sequence accession numbers
All newly derived mosquito Plasmodiidae sequences have been submitted to GenBank with accession numbers MK502145 to MK502166.
Statistical analysis
A generalized linear model (GLM) with binomial distribution and logit link function was used to test the difference in trapping efficacy of the BG‐Sentinel trap and the Suna trap, as well as the attractiveness of different host odours to different mosquito species. Differences in treatments were expressed as the number of mosquitoes caught (per species) in one trap divided by the total number of mosquitoes (per species) trapped during each trapping night (Busula et al., 2015). Effects of location, day, temperature, humidity, CO2 treatment and mosquito species, as well as their two‐way interactions, on the number of mosquitoes caught were fitted in the GLM and non‐significant factors were removed. Models were compared by the corrected Akaike's information criterion. P < 0.05 was considered statistically significant. All statistical analyses were performed using spss, version 20 (IBM Corp., Armonk, NY, U.S.A.).
Results
Mosquito host preference and identification
Mosquito trapping
In total, 5145 Anopheles mosquitoes were caught during the study period, the vast majority of which (n = 5002) were identified as Anopheles obscurus. The remaining Anopheles mosquitoes were identified as Anopheles paludis, Anopheles moucheti, Anopheles ziemanni, Anopheles gambiae s.l. and Anopheles nili (see Supporting information, Table S2). Anopheles moucheti (n = 4), An. nili (n = 6) and An. gambiae s.l. (n = 21) are known vectors of human Plasmodia spp. (Wondji et al., 2002; Cohuet et al., 2004; Sinka et al., 2012) and represented 0.6% of all anophelines caught in the present study. Other mosquito species consisted of 6097 Mansonia africana, 1742 Culex spp., 549 Coquillettidia spp. and 68 Aedes spp.
The BG‐Sentinel trap caught more mosquitoes per trap (mean ± SE: 43.89 ± 10.34) than the Suna trap (25.17 ± 6.11); however, these means were not significantly different after inclusion of the location effect in the GLM (GLM, P = 0.204 for trap and P < 0.001 for location) (Fig. 1). The mean number of Anopheles mosquitoes per trap per day was 1.50 ± 0.59 for the BG‐Sentinel trap and 1.47 ± 0.5 for the Suna trap. Because the mosquito trap catches were not significantly different between the two traps and the Suna trap is more weather resistant than the BG‐Sentinel trap, the Suna trap was chosen for further experiments.
Figure 1.
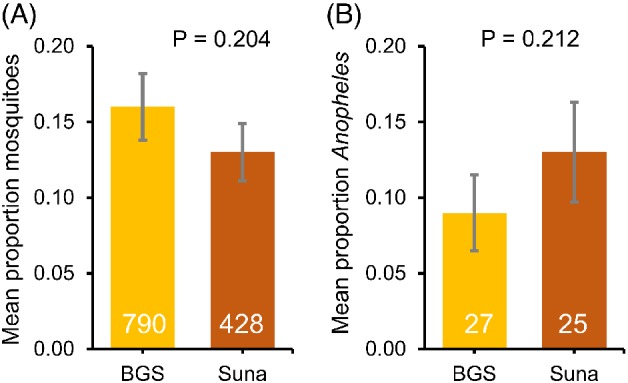
Trapping efficiency of two odour‐baited mosquito traps. Back‐transformed mean proportion [generalized linear model (GLM)] of caught mosquitoes per trap using the BG‐Sentinel (BGS) trap (n = 15) and the Suna trap (n = 15) baited with the five‐component odour blend odour blend (Menger et al., 2014) and CO2. Numbers in the bars indicate the total number of mosquito spp. (A) and Anopheles spp. (B) trapped. Error bars represent the SEM. Location effect was significant for both total mosquito spp. and total Anopheles spp. and included in the GLM (P < 0.001). [Colour figure can be viewed at http://wileyonlinelibrary.com].
Host preference of different mosquito species
The majority of the species caught during 35 trapping nights were Anopheles spp. (n = 3347), Mansonia spp. (n = 3542), Culex spp. (n = 1645) and Coquillettidia spp. (n = 541).
Mosquito catches were significantly affected by trap location (GLM, P < 0.001) and odour treatments (P < 0.001). In addition, there was a significant interaction between the type of odour bait used and mosquito species (GLM, P < 0.038), indicating that the odour baits differentially attracted the mosquito species present in the area. At some locations, certain mosquito species were more abundant than others, as indicated by an interaction between mosquito species and location in the GLM (P < 0.001). The significant interactions between mosquito species, odour and location indicated that different mosquito species respond differently to a variation of odours and are more abundant at different locations.
Significantly more Anopheles mosquitoes were caught in the presence of chimpanzee, human, or cow odours compared with traps baited with CO2 alone or without any bait (P < 0.001, GLM) (Fig. 2 and Table 1; see also Supporting information, Table S3 and Figure S1). However, Anopheles mosquitoes (97% of which were identified as An. obscurus) did not show a specific host preference for chimpanzee (mean 29.9 ± 7.6), cow (28.3 ± 8.9) or human (27.0 ± 9.4) odours (P > 0.05, GLM) (Fig. 2 and Table 1; see also Supporting information, Table S3 and Figure S1).
Figure 2.
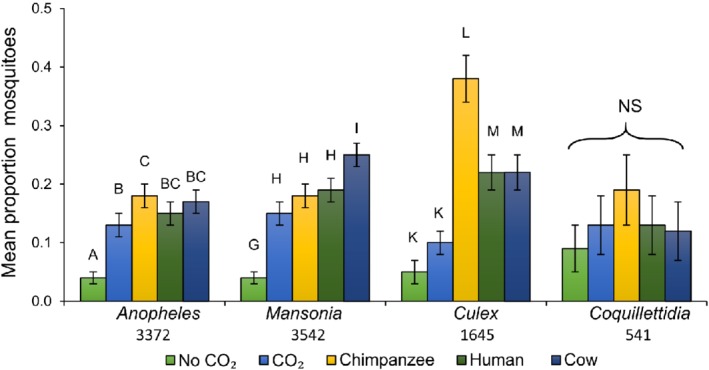
Attraction of mosquito species to different host odours trapped with odour baited Suna traps. Back‐transformed mean proportion [generalized linear model (GLM)] of mosquito genera caught using different host odour treatments: no odours, CO2 only, chimpanzee odours with CO2, human odours with CO2 and cow odours with CO2. Error bars represent the SEM. Numbers below the bars indicate the total number of mosquitoes caught per species (n = 35 trapping nights). Different uppercase letters indicate significant differences between odour baits within each genus (GLM with least significant difference post‐hoc test, P < 0.05). NS, non‐significant (P > 0.05). [Colour figure can be viewed at http://wileyonlinelibrary.com].
Table 1.
Mean number of trapped mosquitoes per night per odour bait.
| Odour bait (mean ± SE) | |||||
|---|---|---|---|---|---|
| Mosquito species | No CO2 | CO2 | Chimpanzee + CO2 | Human + CO2 | Cow + CO2 |
| Anopheles | 5.7 ± 2.7a | 21.6 ± 7.0b | 29.9 ± 7.6c | 27.0 ± 9.4bc | 28.3 ± 8.9bc |
| Mansonia | 10.4 ± 3.0g | 20.9 ± 6.5h | 25.1 ± 4.7h | 27.6 ± 8.5h | 34.1 ± 8.5i |
| Culex | 3.1 ± 1.4k | 6.5 ± 1.5k | 20.4 ± 4.9l | 12.8 ± 3.6m | 12.0 ± 2.4m |
| Coquillettidia | 2.1 ± 0.8 | 2.8 ± 1.0 | 5.0 ± 1.6 | 4.2 ± 2.3 | 3.9 ± 1.2 |
There were 35 trapping nights in total. Different superscript lowercase letters indicate significant differences between odour baits for each species (generalized linear model with least significant difference post‐hoc test, P < 0.05). No significant differences were found for Coquillettidia.
The Mansonia species caught during the present study were morphologically identified as Mansonia africana. Unlike An. obscurus, M. africana exhibited a preference for cow odour (P < 0.05, GLM) (Fig. 2 and Table 1; see also Supporting information, Table S3). Traps baited with cow odour caught an average of 34.1 ± 8.5 M. africana, which was more than three times higher than traps with no bait (10.4 ± 3.0, P < 0.001, GLM). No significant differences were found between chimpanzee or human odours (25.1 ± 4.7, 27.6 ± 8.5, respectively). Traps without bait attracted significantly fewer Mansonia species than with bait (P < 0.05, GLM) (Fig. 2 and Table 1; see also Supporting information, Table S3 and Figure S1).
The Culex species caught in the present study were morphologically identified as Culex decens. Traps baited with chimpanzee odour caught approximately two‐fold more Culex decens (20.4 ± 4.9) than traps baited with cow (12.0 ± 2.4, P = 0.002, GLM) or human odour (12.8 ± 3.6, P = 0.002, GLM). Adding CO2 to a Suna trap did not result in a significant increase in mosquito catches compared with traps with no bait (P = 0.083, GLM) (Fig. 2 and Table 1).
No significant differences in mosquito attraction towards different host odours were observed for Coquillettidia species (n = 541, P > 0.05) (Fig. 2 and Table 1; see also Supporting information, Table S3 and Figure S1).
Plasmodium detection
In total, 84 chimpanzees (Pan troglodytes) were screened for Plasmodium species. Despite the fact that other groups have detected ape Plasmodium species at this field site (Pacheco et al., 2013), no ape Plasmodium species were detected during the course of the present study. However, two chimpanzees were found to be infected with the human parasite P. falciparum (Fig. 3).
Figure 3.
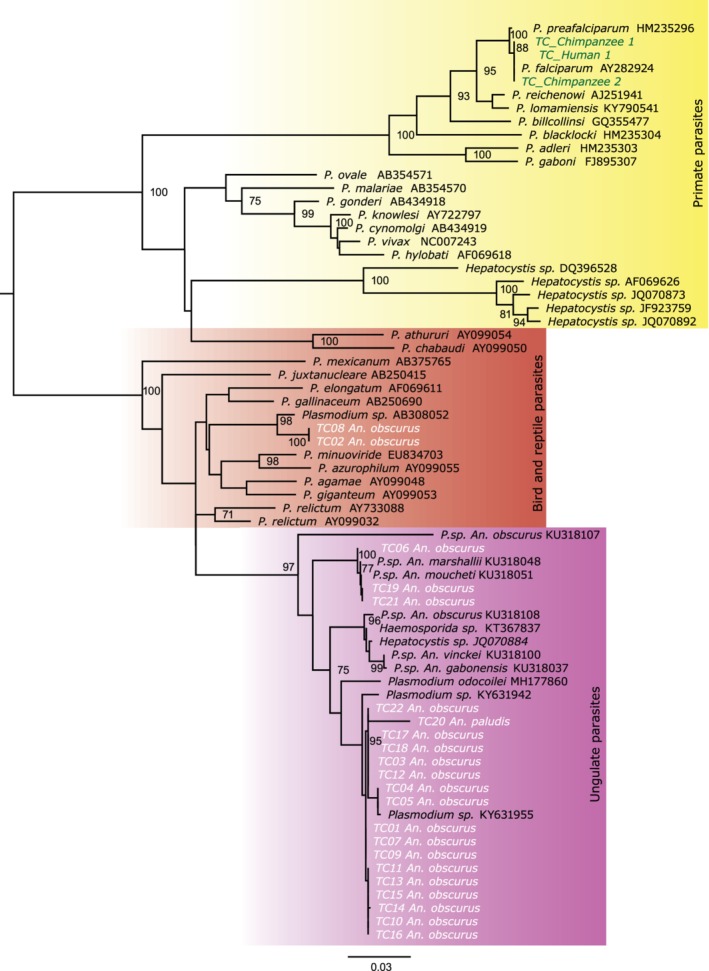
Evolutionary relationships of Plasmodium parasite sequences from chimpanzees and humans. A maximum likelihood tree of mitochondrial cytochrome b (cytB) sequences (956 bp) is shown. Sequences in green represent the Plasmodium sequences found in chimpanzee (Chimpanzee 1 and 2) and human (Human 1) blood samples. Sequences in white represent Plasmodiidae sequences obtained from Anopheles mosquitoes. Black sequences represent Plasmodium reference sequences. Coloured blocks indicate primate (yellow), bird and reptile (brown), and ungulate (purple) reference sequences. Bootstrap values (≥ 70%) are shown above and below the branch nodes. The scale bar represents 0.03 nucleotide substitutions per site. [Colour figure can be viewed at http://wileyonlinelibrary.com].
The Anopheles mosquitoes (n = 5145) and the preserved blood meals from Mansonia species (n = 144) collected using odour‐baited traps were also screened for Plasmodium spp. One P. falciparum parasite was found in the human blood meal obtained from a M. africana mosquito (Fig. 3). Moreover, Plasmodiidae species were amplified from 21 An. obscurus mosquitoes (0.4%) and one An. paludis mosquito (4.3%) (Fig. 3). Consistent with the lack of ape Plasmodium parasites in the chimpanzee population, phylogenetic analysis revealed that none of the Plasmodiidae species detected were of ape origin. Most of the Plasmodiidae sequences (n = 17) formed a single clade distinct from characterized Plasmodium species (Fig. 3). They did cluster with a Plasmodium spp. from an African buffalo (Syncerus caffer) from Gabon and a Plasmodium spp. from a marshbuck (Tragelaphus spekii) from Gabon (Bitome‐Essono et al., 2017). Three sequences (TC06, TC021 and TC019) clustered with a Plasmodium spp. that has previously been amplified from An. moucheti and Anopheles marshalii mosquitoes in Gabon (Boundenga et al., 2016). Two Plasmodiidae sequences (TC02 and TC08) clustered with a Plasmodium spp. from a Coquillitidia mosquito from Japan (Ejiri et al., 2008). In addition, these two Plasmodiidae sequences belong to a group of Plasmodium spp. that are found in birds (Ricklefs & Outlaw 2010).
Discussion
When sampling mosquitoes at a chimpanzee rehabilitation site, situated within a natural chimpanzee habitat where wild mosquitoes would feed on the chimpanzees being rehabilitated, it was found that mosquito species exhibited different host preferences. Moreover, the location of the trap had a significant effect on the mosquito catches, as seen in other field studies on mosquito host preference (Hiscox et al., 2014; Pombi et al., 2014). Most of the mosquito species caught during the present study, including An. obscurus and M. africana, showed a generalistic host preference and were attracted to all of the host species tested. Interestingly, a Culex species belonging to the Cx. decens complex showed a much higher attraction towards chimpanzee odours than to human or cow odours. The blood‐feeding behaviour of mosquitoes is highly plastic and the adaptation of mosquitoes to available host species could have implications for pathogen transmission (Chaves et al., 2010; Takken & Verhulst 2013). Studies have shown that Cx. decens feeds on both birds and bats (Boreham & Snow 1973; Quan et al., 2010). Whether Cx. decens has developed a specialised host preference for chimpanzees remains to be investigated. The specialisation of mosquitoes could turn a generalist mosquito into a more dangerous anthropophilic vector of human infections, as seen for Aedes aegypti, which is now a dominant vector of Zika virus and dengue virus (Yakob et al., 2010; Scott & Takken 2012; Brown et al., 2014).
Mansonia mosquitoes are aggressive biting mosquitoes and are associated with Rift Valley fever virus, West Nile virus and Wucheria bancrofti (Fontenille et al., 1998; Diallo et al., 2005; Ughasi et al., 2012). The Mansonia species caught during the present study were all morphologically identified as M. africana and showed a strong preference for cow odour‐baited mosquito traps. Different studies in the past have observed different host preferences for Mansonia mosquitoes, ranging between anthropophilic, a high preference for cows and a generalistic host preference, respectively (Lefèvre et al., 2009; Busula et al., 2015; Omondi et al., 2015). Although the different results obtained could be a result of the study methodology, Busula et al. (2015) utilized a comparable experimental design to that used in the present study, indicating that, even within a mosquito species, different populations can have different host preferences (Takken & Verhulst 2013).
Anopheles obscurus was the predominant Anopheles species caught during the present study (97.4%). This typical marsh and swamp breeder is an opportunistic species biting both bovine and primate hosts (Boorman and Service, Boorman & Service 1960). Anopheles obscurus has also been associated with feeding on ungulate hosts such as Cephalophus (duikers) species and found to be infected with ungulate Plasmodium spp. (Boundenga et al., 2016; Makanga et al., 2017). Experimental studies have shown that Anopheles mosquitoes with different host preferences are similarly attracted towards non‐human primates and humans (Verhulst et al., 2018). Moreover, a field study by Makanga et al. (2016) identified mosquitoes biting both humans and great apes. The Anopheles mosquitoes in the present study were equally attracted towards chimpanzee and human odours. However, although 21 An. obscurus mosquitoes were infected with a variety of Plasmodiidae species, no primate Plasmodium was detected, which suggests that An. obscurus in not a competent vector. Because of the lack of well‐defined reference sequences, it was impossible to classify the parasites to genus level with any kind of certainty. Therefore, it was decided to assign them to the Plasmodiidae family, which includes both Plasmodium and Hepatocystis. Other mosquito species such as An. gambiae, An. ziemanni, An. nili and An. moucheti caught in the present study are known Plasmodium vectors and may play a role as bridge vectors in the circulation of P. falciparum in this area. Although their abundance was very low, this could still be sufficient to sustain transmission (Homan et al., 2016).
Human P. falciparum was found in a chimpanzee blood sample and a human blood sample from a Mansonia mosquito (Fig. 3), which indicated that chimpanzees can be infected with P. falciparum. Although a chimpanzee to chimpanzee transmission of P. falciparum cannot be ruled out, it is likely that this represents transmission of P. falciparum from humans to chimpanzees, which has previously been observed in apes held in captivity (Krief et al., 2010; Prugnolle et al., 2010; Pacheco et al., 2013; Ngoubangoye et al., 2016). By contrast, P. falciparum has never been detected in wild chimpanzees, bonobos or gorillas, which suggests that there is something unique about the sanctuary environment that facilitates cross‐species transmission. It is possible that the rangers working in the sanctuary harbour subclinical densities of P. falciparum (Rayner et al., 2011; Maselli et al., 2014; Rovira‐Vallbona et al., 2017), which, when picked up by anopheline mosquitoes, can be transmitted to the chimpanzees held in the sanctuary. However, it was not possible to identify bridge vectors for P. falciparum because none of the Anopheles specimens collected were infected with human P. falciparum, although some of the mosquitoes caught at Tchimpounga (An. ziemanni, An. nili, An. moucheti and gambiae s.l.) are known human malaria vectors (Greenwood et al., 2005; Scott & Takken 2012; Sinka et al., 2012). Additional work is needed to determine the factors that facilitate cross species transmission of P. falciparum in sanctuary apes.
Unravelling the trophic behaviour of mosquitoes in remote locations will provide us with valuable information on potential transmission pathways of vector‐borne diseases between non‐human primates. With increasing human activities in natural environments, the risk of zoonoses increases, as seen in the cases of a non‐human primate derived P. vivax infecting a Caucasian man travelling from Africa and the transmission of Plasmodium knowlesi from monkey to humans in South‐East Asia (Cox‐Singh et al., 2008; Singh et al., 2004; Paupy et al., 2013). This is further supported by recent zoonotic transmission of P. simium from simians to humans in the Atlantic Forest of Rio de Janeiro (Brasil et al., 2017). Mosquito plasticity in host preference has recently been shown as a major factor for disease outbreaks (Yakob et al., 2018). In addition, adaptation of arthropod‐borne viruses to new mosquito vectors is a major concern in the spread of vector‐borne diseases and has already caused outbreaks of chikungunya virus and West‐Nile virus (Kilpatrick et al., 2006, Kilpatrick et al., 2008, Tsetsarkin et al., 2007, 2016). The trophic behaviour of mosquitoes was determined in the present study using a variety of methods, including odour‐baited traps, analyses of blood meal and pathogens of mosquitoes. Each of these methods alone would have led to different conclusions, which showed that a combination of methods is required to be able to fully understand the behaviour of disease vectors. The present study provides evidence that the majority of mosquito species collected near wild apes are attracted to odours of multiple different species, including chimpanzees and humans, and may thus serve as bridge vectors for ape pathogens.
The identification of P. falciparum in both humans and chimpanzees suggests that there is active circulation of these parasites transmitted by anopheline mosquitoes, although it is unlikely that An. obscurus is part of this transmission cycle given that, regardless of their high abundance, no primate Plasmodium positive specimens were found. Primary human Plasmodium vectors such as An. gambiae s.l., An. nili and An. moucheti, were collected during this study period, which could have played a role as bridge vectors for the circulation of P. falciparum.
Supporting information
Figure S1. Attraction of mosquito species to different host odours trapped with odour‐baited Suna traps.
Table S1. Plasmodium cytB sequences used in the phylogenetic analyses.
Table S2. Samples collected and prevalence of Plasmodium.
Table S3. Pairwise comparisons of mosquito trap catches with different odour baits.
Acknowledgements
The research was supported by a grant from the Earth and Life Sciences Foundation (Veni‐ALW) of the Netherlands Organisation for Scientific Research (NWO, 863.13.012), as well as grants from the NIH (R01 AI097137, R01 AI091595). We thank the Jane Goodall institute and the Tchimpounga Chimpanzee Rehabilitation Centre in the Republic of Congo for providing access to conduct our research and to collect all samples. Special gratitude goes to R. Attencia and D. Cox, as well as the TC caretakers, for their assistance at the Jane Goodall Institute. We thank J. Rayner for help in writing the project proposal and C. J. M. Koenraadt for assistance in setting up the field experiments. We thank A. Cornel from UC Davis for help with the mosquito identification. We are thankful to Weimin Liu and Yinging Li for technical assistance. The authors declare that they have no conflicts of interest.
References
- Bitome‐Essono, P.Y. , Ollomo, B. , Arnathau, C. et al (2017) Tracking zoonotic pathogens using blood‐sucking flies as ‘flying syringes’. eLife, 6, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorman, J.P. & Service, M.W. (1960) Some records of mosquitoes (Culicidae, Diptera) from The Niger Delta area, Southern Nigeria. West African Journal of Medicine, 9, 67–72. [PubMed] [Google Scholar]
- Boreham, P.F.L. & Snow, W.F. (1973) Further information on the food sources of Culex (Culex) decens (Theo. Dipt., Culicidae). Transactions of the Royal Society of Tropical Medicine and Hygiene, 67, 724–725. [DOI] [PubMed] [Google Scholar]
- Boundenga, L. , Makanga, B. , Ollomo, B. et al (2016) Haemosporidian parasites of antelopes and other vertebrates from Gabon, Central Africa. PLoS ONE, 11, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasil, P. , Zalis, M.G. , de Pina‐Costa, A. , Siqueira, A.M. , Júnior, C. et al (2017) Outbreak of human malaria caused by Plasmodium simium in the Atlantic Forest in Rio de Janeiro: a molecular epidemiological investigation. The Lancet Global Health, 5, 1038–1045. [DOI] [PubMed] [Google Scholar]
- Brown, J.E. , Evans, B.R. , Zheng, W. et al (2014) Human impacts have shaped historical and recent evolution in Aedes aegypti, the dengue and yellow fever mosquito. Evolution (N. Y)., 68, 514–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busula, A.O. , Takken, W. , Loy, D.E. , Hahn, B.H. , Mukabana, W.R. & Verhulst, N.O. (2015) Mosquito host preferences affect their response to synthetic and natural odour blends. Malaria Journal, 14, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busula, A.O. , Takken, W. , de Boer, J.G. , Mukabana, W.R. & Verhulst, N.O. (2017) Variation in host preferences of malaria mosquitoes is mediated by skin bacterial volatiles. Medical and Veterinary Entomology, 31, 320–326. [DOI] [PubMed] [Google Scholar]
- Chaves, L.F. , Harrington, L.C. , Keogh, C.L. , Nguyen, A.M. & Kitron, U.D. (2010) Blood feeding patterns of mosquitoes: random or structured? Frontiers in Zoology, 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohuet, A. , Simard, F. , Wondji, C.S. , Antonio‐Nkondjio, C. , Awono‐Ambene, P. & Fontenille, D. (2004) High malaria transmission intensity due to Anopheles funestus (Diptera: Culicidae) in a village of savannah‐forest transition area in Cameroon. Journal of Medical Entomology, 41, 901–905. [DOI] [PubMed] [Google Scholar]
- Cox‐Singh, J. , Davis, T.M. , Lee, K.S. et al (2008) Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clinical Infectious Diseases, 46, 165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba, D. , Taboada, G.L. , Doallo, R. & Posada, D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods, 9, 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Délicat‐Loembet, L. , Rougeron, V. , Ollomo, B. et al (2015) No evidence for ape Plasmodium infections in humans in Gabon. PLoS ONE, 10, e0126933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diallo, M. , Nabeth, P. , Ba, K. et al (2005) Mosquito vectors of the 1998‐1999 outbreak of Rift Valley fever and other arboviruses (Bagaza, Sanar, Wesselsbron and West Nile) in Mauritania and Senegal. Medical and Veterinary Entomology, 19, 119–126. [DOI] [PubMed] [Google Scholar]
- Ejiri, H. , Sato, Y. , Sasaki, E. et al (2008) Detection of avian Plasmodium spp. DNA sequences from mosquitoes captured in Minami Daito Island of Japan. Journal of Veterinary Medical Science, 70, 1205–1210. [DOI] [PubMed] [Google Scholar]
- Fauci, A.S. & Morens, D.M. (2016) Zika virus in the Americas—yet another arbovirus threat. New England Journal of Medicine, 374, 601–604. [DOI] [PubMed] [Google Scholar]
- Fontenille, D. , Traore‐Lamizana, M. , Diallo, M. , Thonnon, J. , Digoutte, J.P. & Zeller, H.G. (1998) New vectors of Rift Valley fever in West Africa. Emerging Infectious Diseases, 4, 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood, B.M. , Bojang, K. , Whitty, C.J.M. & Targett, G.A.T. (2005) Malaria. Lancet, 365, 1487–1498. [DOI] [PubMed] [Google Scholar]
- Gillies, M.T. & Coetzee, M. (1987) A supplement to the Anophelinae of Africa South of the Sahara. Publications of the South African Institute for Medical Research, 55, 148. [Google Scholar]
- Guindon, S. , Dufayard, J.‐F. , Lefort, V. , Anisimova, M. , Hordijk, W. & Gascuel, O. (2010) New algorithms and methods to estimate maximum‐likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology, 59, 307–321. [DOI] [PubMed] [Google Scholar]
- Harrington, L.C. , Edman, J.D. & Scott, T.W. (2001) Why do female Aedes aegypti (Diptera: Culicidae) feed preferentially and frequently on human blood? Journal of Medical Entomology, 38, 411–422. [DOI] [PubMed] [Google Scholar]
- Hiscox, A. , Otieno, B. , Kibet, A. et al (2014) Development and optimization of the Suna trap as a tool for mosquito monitoring and control. Malaria Journal, 13, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highton R.B. (1983) Taxonomic keys for the identification of the afrotropical mosquitoes. Unpublished, 1983, 1–85. [Google Scholar]
- Homan, T. , Hiscox, A. , Mweresa, C.K. et al (2016) The effect of mass mosquito trapping on malaria transmission and disease burden (SolarMal): a stepped‐wedge cluster‐randomised trial. Lancet, 388, 207–211. [DOI] [PubMed] [Google Scholar]
- Jones, K.E. , Patel, N.G. , Levy, M.A. et al (2008) Global trends in emerging infectious diseases. Nature, 451, 990–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick, A.M. , Kramer, L.D. , Jones, M.J. , Marra, P.P. & Daszak, P . (2006) West Nile virus epidemics in North America are driven by shifts in mosquito feeding behavior. PLoS Biology, 4, e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick, A.M. , Meola, M.A. , Moudy, R.M. & Kramer, L.D. (2008) Temperature, viral genetics, and the transmission of West Nile virus by Culex pipiens mosquitoes. PLoS Pathogens, 4, e1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krief, S. , Escalante, A.a. , Pacheco, M.A. et al (2010) On the diversity of malaria parasites in African apes and the origin of Plasmodium falciparum from bonobos. PLoS Pathogens, 6, e1000765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts, L. , Scott, T.W. & Gubler, D.J. (2010) Consequences of the expanding global distribution of Aedes albopictus for dengue virus transmission. PLoS Neglected Tropical Diseases, 4, e646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefèvre, T. , Gouagna, L. , Dabiré, K.R. et al (2009) Beyond nature and nurture: phenotypic plasticity in blood‐feeding behavior of Anopheles gambiae s. s. when humans are not readily accessible. The American Journal of Tropical Medicine and Hygiene, 81, 1023–1029. [DOI] [PubMed] [Google Scholar]
- Liu, W. , Li, Y. , Learn, G.H. et al (2010) Origin of the human malaria parasite in gorillas. Nature, 467, 420–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Li, Y. , Shaw, K.S. et al (2014) African origin of the malaria parasite Plasmodium vivax . Nature Communications, 5, 3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Sundararaman, S.A. , Loy, D.E. et al (2016) Multigenomic delineation of Plasmodium species of the Laverania subgenus infecting wild‐living chimpanzees and gorillas. Genome Biology and Evolution, 8, 1929–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Sherrill‐Mix, S. , Learn, G.H. et al (2017) Wild bonobos host geographically restricted malaria parasites including a putative new Laverania species. Nature Communications, 8, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy, D.E. , Liu, W. , Li, Y. et al (2016) Out of Africa: origins and evolution of the human malaria parasites Plasmodium falciparum and Plasmodium vivax . International Journal for Parasitology, 47, 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy, D.E. , Plenderleith, L.J. , Sundararaman, S.A. et al (2018a) Evolutionary history of human Plasmodium vivax revealed by genome‐wide analyses of related ape parasites. Proceedings of the National Academy of Sciences, 115, E8450–E8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy, D.E. , Rubel, M.A. , Avitto, A.N. et al (2018b) Investigating zoonotic infection barriers to ape Plasmodium parasites using faecal DNA analysis. International Journal for Parasitology, 48, 531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makanga, B. , Yangari, P. , Rahola, N. et al (2016) Ape malaria transmission and potential for ape‐to‐human transfers in Africa. Proceedings of the National Academy of Sciences, 113, 5329–5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makanga, B. , Costantini, C. , Rahola, N. et al (2017) ‘Show me which parasites you carry and I will tell you what you eat’, or how to infer the trophic behavior of hematophagous arthropods feeding on wildlife. Ecololgy and Evolution, 7, 7578–7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maselli, L.M. , Levy, D. , Laporta, G.Z. et al (2014) Detection of Plasmodium falciparum and Plasmodium vivax subclinical infection in non‐endemic region: implications for blood transfusion and malaria epidemiology. Malaria Journal, 13, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menger, D. , van Loon, J. & Takken, W. (2014) Assessing the efficacy of candidate mosquito repellents against the background of an attractive source that mimics a human host. Medical and Veterinary Entomology, 28, 407–413. [DOI] [PubMed] [Google Scholar]
- Mweresa, C.K. , Omusula, P. , Otieno, B. et al (2014) Molasses as a source of carbon dioxide for attracting the malaria mosquitoes Anopheles gambiae and Anopheles funestus. Malaria Journal, 13, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngoubangoye, B. , Boundenga, L. , Arnathau, C. et al (2016) The host specificity of ape malaria parasites can be broken in confined environments. International Journal for Parasitology, 46, 737–744. [DOI] [PubMed] [Google Scholar]
- Ndo, C. , Antonio‐Nkondjio, C. , Cohuet, A . et al (2010) Population genetic structure of the malaria vector Anopheles nili in sub‐Saharan Africa. Malaria journal, 9, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omondi, D. , Masiga, D.K. , Ajamma, Y.U. & Fielding, B.C. (2015) Unraveling host‐vector‐arbovirus interactions by two‐gene high resolution melting mosquito bloodmeal analysis in a Kenyan wildlife‐livestock interface. PLoS ONE, 10, e0134375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco, M.A. , Cranfield, M. , Cameron, K. & Escalante, A.a. (2013) Malarial parasite diversity in chimpanzees: the value of comparative approaches to ascertain the evolution of Plasmodium falciparum antigens. Malaria Journal, 12, 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paupy, C. , Makanga, B. , Ollomo, B. et al (2013) Anopheles moucheti and Anopheles vinckei are candidate vectors of ape Plasmodium parasites, including Plasmodium praefalciparum in Gabon. PLoS ONE, 8, e57294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombi, M. , Jacobs, F. , Verhulst, N.O. , Caputo, B. , Torre, A.d. & Takken, W. (2014) Field evaluation of a novel synthetic odour blend and of the synergistic role of carbon dioxide for sampling host‐seeking Aedes albopictus adults in Rome, Italy. Parasites and Vectors, 7, 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prugnolle, F. , Durand, P. , Neel, C. et al (2010) African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum . Proceedings of the National Academy of Sciences, 107, 1458–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, Y.T. , Smallegange, R.C. , ter Braak, C.J.F. et al.et al (2007) Attractiveness of MM‐X traps baited with human or synthetic odor to mosquitoes (Diptera: Culicidae) in the Gambia. Journal of Medical Entomology, 44, 970–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan, P.L. , Junglen, S. , Tashmukhamedova, A. et al (2010) Moussa virus: a new member of the Rhabdoviridae family isolated from Culex decens mosquitoes in Cote d'Ivoire. Virus Research, 147, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner, J.C. , Liu, W. , Peeters, M. , Sharp, P.M. & Hahn, B.H. (2011) A plethora of Plasmodium species in wild apes: a source of human infection? Trends in Parasitology, 27, 222–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklefs, R.E. & Outlaw, D.C. (2010) A molecular clock for malaria parasites. Science, 329, 226–229. [DOI] [PubMed] [Google Scholar]
- Rovira‐Vallbona, E. , Contreras‐Mancilla, J.J. , Ramirez, R. et al (2017) Predominance of asymptomatic and sub‐microscopic infections characterizes the Plasmodium gametocyte reservoir in the Peruvian Amazon. PLoS Neglected Tropical Diseases, 11, e0005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallegange, R.C. , Schmied, W.H. , van Roey, K.J. et al (2010) Sugar‐fermenting yeast as an organic source of carbon dioxide to attract the malaria mosquito Anopheles gambiae. Malaria Journal, 9, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmied, W.H. , Takken, W. , Killeen, G.F. , Knols, B.G.J. & Smallegange, R.C. (2008) Evaluation of two counterflow traps for testing behaviour‐mediating compounds for the malaria vector Anopheles gambiae s.s. under semi‐field conditions in Tanzania. Malaria Journal, 7, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, T.W. & Takken, W. (2012) Feeding strategies of anthropophilic mosquitoes result in increased risk of pathogen transmission. Trends in Parasitology, 28, 114–121. [DOI] [PubMed] [Google Scholar]
- Singh, B. , Sung, L.K. , Matusop, A. et al (2004) A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet, 363, 1017–1024. [DOI] [PubMed] [Google Scholar]
- Sinka, M.E. , Bangs, M.J. , Manguin, S. et al (2012) A global map of dominant malaria vectors. Parasites and Vectors, 5, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararaman, S.A. , Liu, W. , Keele, B.F. et al (2013) Plasmodium falciparum‐like parasites infecting wild apes in southern Cameroon do not represent a recurrent source of human malaria. Proceedings of the National Academy of Sciences, 110, 7020–7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken, W. & Knols, B.G.J. (1999) Odor‐mediated behaviour of Afrotropcial malaria mosquitoes. Annual Review of Entomology, 44, 131–157. [DOI] [PubMed] [Google Scholar]
- Takken, W. & Verhulst, N.O. (2013) Host preferences of blood‐feeding mosquitoes. Annual Review of Entomology, 58, 433–453. [DOI] [PubMed] [Google Scholar]
- Takken, W. & Verhulst, N.O. (2017) Chemical signaling in mosquito–host interactions: the role of human skin microbiota. Current Opinion in Insect Science, 20, 68–74. [DOI] [PubMed] [Google Scholar]
- Tsetsarkin, K.A. , Vanlandingham, D.L. , McGee, C.E. & Higgs, S. (2007) A single mutation in Chikungunya virus affects vector specificity and epidemic potential. PLoS Pathogens, 3, 1895–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin, K.A. , Chen, R. & Weaver, S.C. (2016) Interspecies transmission and chikungunya virus emergence. Current Opinion in Virology, 16, 143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ughasi, J. , Bekard, H.E. , Coulibaly, M. et al (2012) Mansonia africana and Mansonia uniformis are vectors in the transmission of Wuchereria bancrofti lymphatic filariasis in Ghana. Parasites and Vectors, 5, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst, N.O. , Smallegange, R.C. & Takken, W. (2012) Mosquitoes as potential bridge vectors of malaria parasites from non‐human primates to humans. Frontiers in Physiology, 3, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst, N.O. , Umanets, A. , Weldegergis, B.T. et al (2018) Do apes smell like humans? The role of skin bacteria and volatiles of primates in mosquito host selection. Journal of Experimental Biology, 221, 22. [DOI] [PubMed] [Google Scholar]
- Wondji, C.S. , Antonio‐Nkondjio, C. , Meunier, J.‐Y. et al (2002) High malaria transmission intensity in a village close to Yaounde, the capital city of Cameroon. Journal of Medical Entomology, 39, 350–355. [DOI] [PubMed] [Google Scholar]
- World Health Organization (2018). World Malaria Report 2018. Retrieved from https://www.who.int/malaria/publications/world-malaria-report-2018/en/ [accessed on 19 November 2018].
- Yakob, L. , Bonsall, M.B. & Yan, G. (2010) Modelling knowlesi malaria transmission in humans: vector preference and host competence. Malaria Journal, 9, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakob, L. , Lloyd, A.L. , Kao, R.R. et al (2018) Plasmodium knowlesi invasion following spread by infected mosquitoes, macaques and humans. Parasitology, 145, 101–110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Attraction of mosquito species to different host odours trapped with odour‐baited Suna traps.
Table S1. Plasmodium cytB sequences used in the phylogenetic analyses.
Table S2. Samples collected and prevalence of Plasmodium.
Table S3. Pairwise comparisons of mosquito trap catches with different odour baits.


