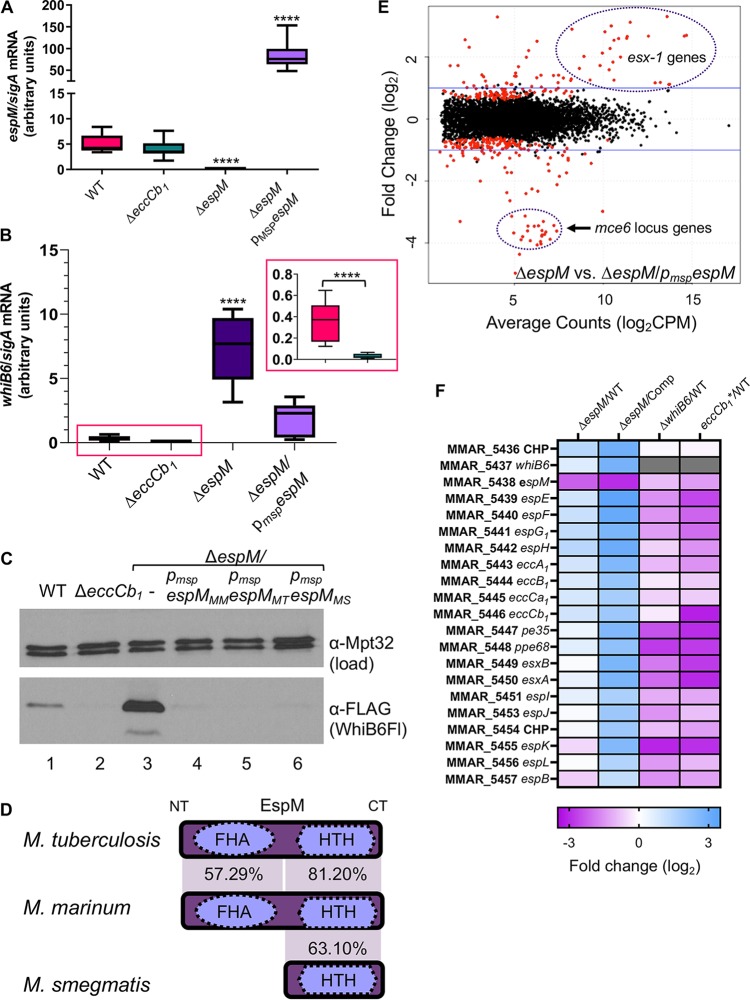
FIG 2.
EspM is a conserved regulator of whiB6 and esx-1 gene expression. (A) qRT-PCR measuring the levels of espM expression relative to sigA expression. A one-way ordinary analysis of variance (ANOVA) (P < 0.0001), followed by a Dunnett’s multiple-comparison test relative to the WT strain, was performed. ****, P < 0.0001. (B) qRT-PCR measuring the levels of whiB6 expression relative to sigA expression. A one-way ordinary ANOVA (P < 0.0001), followed by a Sidak’s multiple-comparison test relative to the WT strain, was performed. ****, P < 0.0001. The inset shows just the comparison between the WT and ΔeccCb1 strains. A Student’s unpaired, two-tailed t test was used to define the significance of the results of the comparisons between the two strains. For panels A and B, the data represent averages of results from at least three biological replicates, each performed in technical triplicate. (C) Western blot analysis of 10 μg of protein per lane. Anti-Mpt32 was used as the loading control. All M. marinum strains indicated in this panel contained a C-terminal FLAG epitope tag on the whiB6 gene. Samples were resolved on an 18% Tris-glycine gel. The Western blot shown is representative of at least three independent biological replicates. All strains indicated in panel D contained a C-terminal epitope tag on the whiB6 gene. (D) Conservation of the EspM proteins (percent identity at the amino acid level) from M. tuberculosis, M. marinum, and M. smegmatis. (E) Scatterplot of genes differentially expressed in the ΔespM strain versus the ΔespM/pmspespM complemented strain. Genes indicated in red had a q value of <0.05. Regions enriched with the esx-1 locus or mce6 locus are highlighted. Full gene lists are available in Table S3. (F) Heat map of esx-1 locus genes that are significantly differentially regulated in the ΔespM strain versus the ΔespM/pmspespM complemented strain compared to genes expressed in the ΔespM, ΔwhiB6, or eccCb1 mutant strains relative to the WT strain.