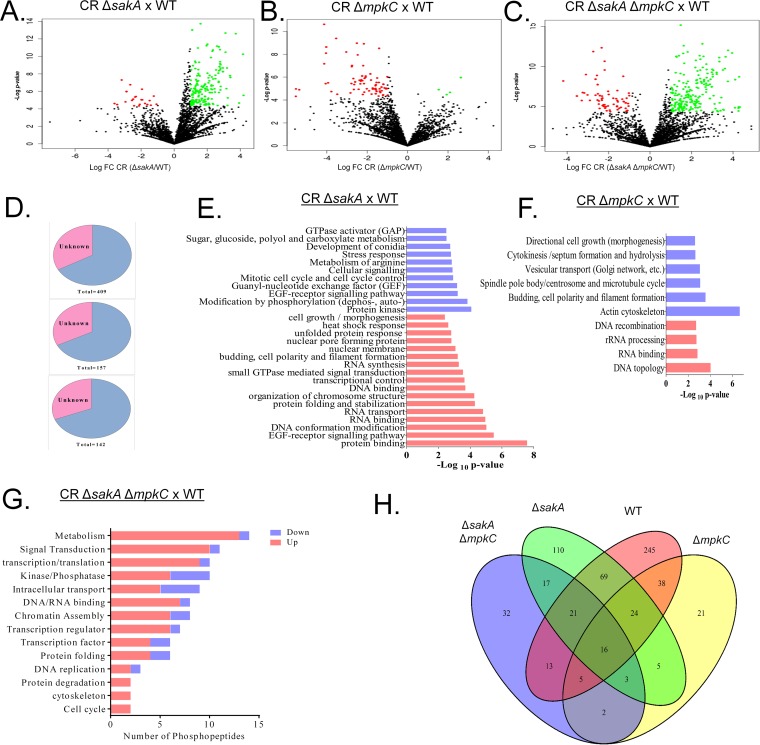
FIG 4.
Phosphorylation profile of proteins from A. fumigatus and MAPK mutants during incubation with Congo red (CR 10 min). (A to C) Volcano plots of total phosphopeptides identified in ΔsakA mutant versus wild type (WT), ΔmpkC mutant versus WT, and the double ΔsakA ΔmpkC mutant versus WT, respectively. (D) Percentage of proteins with unknown and known function in all three phosphoproteomes ΔsakA (top); ΔmkpC (middle); and ΔsakA ΔmpkC (bottom). All of them showed more than 60% characterized proteins. (E to G) Functional categorization of approximately 75% of proteins with known function differentially phosphorylated in ΔsakA, ΔmpkC, and ΔsakA ΔmpkC strains during CR stress. (H) Venn diagram of total differentially phosphorylated proteins (n = 621) representing the number of common phosphoproteins between strains in each intersection (each strain has a set of differentially phosphorylated proteins after CR treatment compared to its untreated control). Thus, the WT strain has a total set of 431 phosphoproteins from which a unique set of 245 is not shared with any other condition, 69 are shared only with the ΔsakA mutant, 13 are shared only with the ΔsakA ΔmpkC mutant, and 38 are shared only with the ΔmpkC mutant, while 16 phosphoproteins are shared simultaneously by the four strains.