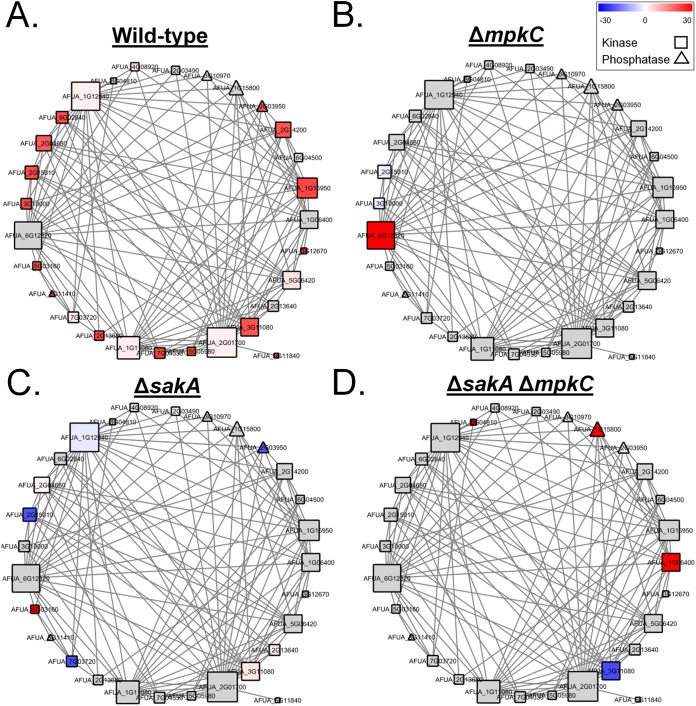
FIG 6.
Subnetwork of functional protein associations based only on the phosphorylation profile of protein kinases and phosphatases during incubation with Congo red (CR 10 min). The subset of differentially phosphorylated kinases and phosphatases from the wild-type (WT), ΔsakA, ΔmpkC, and ΔsakA ΔmpkC strains during CR stress was extracted from the general protein association network. Each node represents a protein that was differentially phosphorylated in at least one strain under CR stress. Node colors are divided into gray nodes (proteins that were not differentially phosphorylated) and heatmap-colored nodes (blue-white-red; proteins which were differentially phosphorylated in each strain in relation to its untreated control). Node shapes represent molecular functions and are divided into squares (kinases) and triangles (phosphatases). Each edge represents a functional protein association retrieved from the STRING server (medium confidence threshold of 0.4 for the interaction score). Node size is representative of the number of edges connected to the node. Note that not all the differentially phosphorylated proteins are present in the network, as for many proteins no functional associations were available. Changes in the color patterns of nodes from panels A to D represent changes in the differential phosphorylation of the proteins comprising the network in WT, ΔsakA, ΔmpkC, and ΔsakA ΔmpkC strains, respectively, during CR stress.